Effects of Mutations in the Phenamacril-Binding Site of Fusarium Myosin-1 on Its Motor Function and Phenamacril Sensitivity
- PMID: 32905433
- PMCID: PMC7469408
- DOI: 10.1021/acsomega.0c02886
Effects of Mutations in the Phenamacril-Binding Site of Fusarium Myosin-1 on Its Motor Function and Phenamacril Sensitivity
Abstract
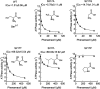
Phenamacril is a Fusarium-specific fungicide used for Fusarium head blight management. The target of phenamacril is FgMyo1, the sole class I myosin in Fusarium graminearum. The point mutation S217L in FgMyo1 is responsible for the high resistance of F. graminearum to phenamacril. Recent structural studies have shown that phenamacril binds to the 50 kDa cleft of the FgMyo1 motor domain, forming extensive interactions, including a hydrogen bond between the cyano group of phenamacril and the hydroxyl group of S217. Here, we produced FgMyo1IQ2, a truncated FgMyo1 composed of the motor domain and two IQ motifs complexed with the F. graminearum calmodulin in insect Sf9 cells. Phenamacril potently inhibited both the basal and the actin-activated ATPase activities of FgMyo1IQ2, with an IC50 in a micromolar range. S217 mutations of FgMyo1IQ2 substantially increased the IC50 of phenamacril. S217T or S217L each increased the IC50 of phenamacril for ∼60-fold, while S217A only increased the IC50 for ∼4-fold. These results indicate that the hydroxyl group of S217 plays an important, but nonessential role in phenamacril binding and that the bulky side chain at the position 217 sterically hinders phenamacril binding. On the other hand, S217P, which might alter the local conformation of the phenamacril-binding site, completely abolished the phenamacril inhibition. Because the cyano group of phenamacril does not form discernible interactions with FgMyo1 other than the nonessential hydrogen bond with the S217 hydroxyl group, we propose the cyano group of phenamacril as a key modification site for the development of novel fungicides.
Copyright © 2020 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Dean R.; Van Kan J. A. L.; Pretorius Z. A.; Hammond-Kosack K. E.; Di Pietro A.; Spanu P. D.; Rudd J. J.; Dickman M.; Kahmann R.; Ellis J.; Foster G. D. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. 10.1111/j.1364-3703.2011.00783.x. - DOI - PMC - PubMed
-
- Li H.; Diao Y.; Wang J.; Chen C.; Ni J.; Zhou M. JS399-19, a new fungicide against wheat scab. Crop Protection 2008, 27, 90–95. 10.1016/j.cropro.2007.04.010. - DOI
LinkOut - more resources
Full Text Sources

