Cholinergic Synaptic Homeostasis Is Tuned by an NFAT-Mediated α7 nAChR-Kv4/Shal Coupled Regulatory System
- PMID: 32905767
- PMCID: PMC7521586
- DOI: 10.1016/j.celrep.2020.108119
Cholinergic Synaptic Homeostasis Is Tuned by an NFAT-Mediated α7 nAChR-Kv4/Shal Coupled Regulatory System
Abstract
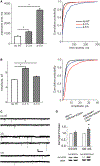
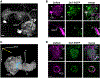
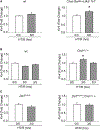
Homeostatic synaptic plasticity (HSP) involves compensatory mechanisms employed by neurons and circuits to preserve signaling when confronted with global changes in activity that may occur during physiological and pathological conditions. Cholinergic neurons, which are especially affected in some pathologies, have recently been shown to exhibit HSP mediated by nicotinic acetylcholine receptors (nAChRs). In Drosophila central neurons, pharmacological blockade of activity induces a homeostatic response mediated by the Drosophila α7 (Dα7) nAChR, which is tuned by a subsequent increase in expression of the voltage-dependent Kv4/Shal channel. Here, we show that an in vivo reduction of cholinergic signaling induces HSP mediated by Dα7 nAChRs, and this upregulation of Dα7 itself is sufficient to trigger transcriptional activation, mediated by nuclear factor of activated T cells (NFAT), of the Kv4/Shal gene, revealing a receptor-ion channel system coupled for homeostatic tuning in cholinergic neurons.
Keywords: Drosophila; K(v)4; NACHO; NFAT; Shal; activity-dependent; cholinergic signaling; homeostatic synaptic plasticity; synaptic homeostasis; α7 nAChR.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

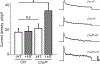

References
-
- Amberg GC, Rossow CF, Navedo MF, and Santana LF (2004). NFATc3 regulates Kv2.1 expression in arterial smooth muscle. J. Biol. Chem 279, 47326–47334. - PubMed
-
- Bischof J, Björklund M, Furger E, Schertel C, Taipale J, and Basler K (2013). A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140, 2434–2442. - PubMed
-
- Daubert EA, and Condron BG (2007). A solid-phase immunostaining protocol for high-resolution imaging of delicate structures in the Drosophila larval central nervous system (CNS). CSH Protoc. 2007, pdb.prot4771. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

