Cytogenetically visible inversions are formed by multiple molecular mechanisms
- PMID: 32906200
- PMCID: PMC7702065
- DOI: 10.1002/humu.24106
Cytogenetically visible inversions are formed by multiple molecular mechanisms
Abstract
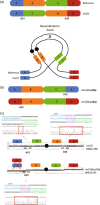
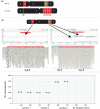
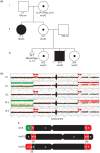
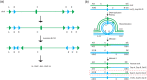
Cytogenetically detected inversions are generally assumed to be copy number and phenotypically neutral events. While nonallelic homologous recombination is thought to play a major role, recent data suggest the involvement of other molecular mechanisms in inversion formation. Using a combination of short-read whole-genome sequencing (WGS), 10X Genomics Chromium WGS, droplet digital polymerase chain reaction and array comparative genomic hybridization we investigated the genomic structure of 18 large unique cytogenetically detected chromosomal inversions and achieved nucleotide resolution of at least one chromosomal inversion junction for 13/18 (72%). Surprisingly, we observed that seemingly copy number neutral inversions can be accompanied by a copy-number gain of up to 350 kb and local genomic complexities (3/18, 17%). In the resolved inversions, the mutational signatures are consistent with nonhomologous end-joining (8/13, 62%) or microhomology-mediated break-induced replication (5/13, 38%). Our study indicates that short-read 30x coverage WGS can detect a substantial fraction of chromosomal inversions. Moreover, replication-based mechanisms are responsible for approximately 38% of those events leading to a significant proportion of inversions that are actually accompanied by additional copy-number variation potentially contributing to the overall phenotypic presentation of those patients.
Keywords: chromosomal inversions; nonallelic homologous recombination; nonhomologous end-joining; recombinant chromosomes; replication-based repair mechanisms; whole-genome sequencing.
© 2020 The Authors. Human Mutation published by Wiley Periodicals LLC.
Conflict of interest statement
James R. Lupski has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered in the Baylor Genetics Laboratory (
Figures



References
-
- Bahrambeigi, V. , Song, X. , Sperle, K. , Beck, C. R. , Hijazi, H. , Grochowski, C. M. , … Lupski, J. R. (2019). Distinct patterns of complex rearrangements and a mutational signature of microhomeology are frequently observed in PLP1 copy number gain structural variants. Genome Medicine, 11(1), 80. - PMC - PubMed

