Genomic Analysis of Selected Maize Landraces from Sahel and Coastal West Africa Reveals Their Variability and Potential for Genetic Enhancement
- PMID: 32906687
- PMCID: PMC7565678
- DOI: 10.3390/genes11091054
Genomic Analysis of Selected Maize Landraces from Sahel and Coastal West Africa Reveals Their Variability and Potential for Genetic Enhancement
Abstract
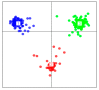
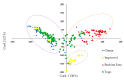
Genetic adaptation of maize to the increasingly unpredictable climatic conditions is an essential prerequisite for achievement of food security and sustainable development goals in sub-Saharan Africa. The landraces of maize; which have not served as sources of improved germplasm; are invaluable sources of novel genetic variability crucial for achieving this objective. The overall goal of this study was to assess the genetic diversity and population structure of a maize panel of 208 accessions; comprising landrace gene pools from Burkina Faso (58), Ghana (43), and Togo (89), together with reference populations (18) from the maize improvement program of the International Institute of Tropical Agriculture (IITA). Genotyping the maize panel with 5974 DArTseq-SNP markers revealed immense genetic diversity indicated by average expected heterozygosity (0.36), observed heterozygosity (0.5), and polymorphic information content (0.29). Model-based population structure; neighbor-joining tree; discriminant analysis of principal component; and principal coordinate analyses all separated the maize panel into three major sub-populations; each capable of providing a wide range of allelic variation. Analysis of molecular variance (AMOVA) showed that 86% of the variation was within individuals; while 14% was attributable to differences among gene pools. The Burkinabe gene pool was strongly differentiated from all the others (genetic differentiation values >0.20), with no gene flow (Nm) to the reference populations (Nm = 0.98). Thus; this gene pool could be a target for novel genetic variation for maize improvement. The results of the present study confirmed the potential of this maize panel as an invaluable genetic resource for future design of association mapping studies to speed-up the introgression of this novel variation into the existing breeding pipelines.
Keywords: DArTseq markers; West Africa; genetic diversity; landraces; maize improvement; population structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Warburton M.L., Reif J.C., Frisch M., Bohn M., Bedoya C., Xia X.C., Crossa J., Franco J., Hoisington D., Pixley K., et al. Genetic diversity in CIMMYT non-temperate maize germplasm: Landraces, open pollinated varieties, and inbred lines. Crop Sci. 2008;48:617–624. doi: 10.2135/cropsci2007.02.0103. - DOI
-
- Byrne P., Richards C., Volk G.M. From wild species to landraces and cultivars. Crop Wild Relatives and their Use in Plant Breeding. In: Byrne P., editor. VolkGM. Colorado State University; Fort Collins, CO, USA: 2020.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

