The Husavirus Posa-Like Viruses in China, and a New Group of Picornavirales
- PMID: 32906743
- PMCID: PMC7551994
- DOI: 10.3390/v12090995
The Husavirus Posa-Like Viruses in China, and a New Group of Picornavirales
Abstract
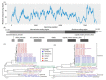
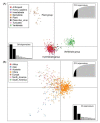
Novel posa-like viral genomes were first identified in swine fecal samples using metagenomics and were designated as unclassified viruses in the order Picornavirales. In the present study, nine husavirus strains were identified in China. Their genomes share 94.1-99.9% similarity, and alignment of these nine husavirus strains identified 697 nucleotide polymorphism sites across their full-length genomes. These nine strains were directly clustered with the Husavirus 1 lineage, and their genomic arrangement showed similar characteristics. These posa-like viruses have undergone a complex evolutionary process, and have a wide geographic distribution, complex host spectrum, deep phylogenetic divergence, and diverse genomic organizations. The clade of posa-like viruses forms a single group, which is evolutionarily distinct from other known families and could represent a distinct family within the Picornavirales. The genomic arrangement of Picornavirales and the new posa-like viruses are different, whereas the posa-like viruses have genomic modules similar to the families Dicistroviridae and Marnaviridae. The present study provides valuable genetic evidence of husaviruses in China, and clarifies the phylogenetic dynamics and the evolutionary characteristics of Picornavirales.
Keywords: Picornavirales; evolution; husavirus; phylogeny; posa-like virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Adams M.J., Lefkowitz E.J., King A.M.Q., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J., Mushegian A., et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017) Arch. Virol. 2017;162:2505–2538. doi: 10.1007/s00705-017-3358-5. - DOI - PubMed
-
- Le Gall O., Christian P., Fauquet C.M., King A.M.Q., Knowles N.J., Nakashima N., Stanway G., Gorbalenya A.E. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 2008;153:715–727. doi: 10.1007/s00705-008-0041-x. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

