Design of Bio-Conjugated Hydrogels for Regenerative Medicine Applications: From Polymer Scaffold to Biomolecule Choice
- PMID: 32906772
- PMCID: PMC7571016
- DOI: 10.3390/molecules25184090
Design of Bio-Conjugated Hydrogels for Regenerative Medicine Applications: From Polymer Scaffold to Biomolecule Choice
Abstract
Bio-conjugated hydrogels merge the functionality of a synthetic network with the activity of a biomolecule, becoming thus an interesting class of materials for a variety of biomedical applications. This combination allows the fine tuning of their functionality and activity, whilst retaining biocompatibility, responsivity and displaying tunable chemical and mechanical properties. A complex scenario of molecular factors and conditions have to be taken into account to ensure the correct functionality of the bio-hydrogel as a scaffold or a delivery system, including the polymer backbone and biomolecule choice, polymerization conditions, architecture and biocompatibility. In this review, we present these key factors and conditions that have to match together to ensure the correct functionality of the bio-conjugated hydrogel. We then present recent examples of bio-conjugated hydrogel systems paving the way for regenerative medicine applications.
Keywords: bio-conjugated hydrogels; biomolecule; polymer scaffold; regenerative medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




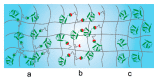



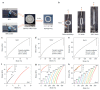

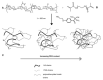


References
-
- Annabi N., Tamayol A., Uquillas J.A., Akbari M., Bertassoni L.E., Cha C., Camci-Unal G., Dokmeci M.R., Peppas N.A., Khademhosseini A. 25th anniversary article: Rational design and applications of hydrogels in regenerative medicine. Adv. Mater. 2014;26:85–124. doi: 10.1002/adma.201303233. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

