Natural display of nuclear-encoded RNA on the cell surface and its impact on cell interaction
- PMID: 32907628
- PMCID: PMC7488101
- DOI: 10.1186/s13059-020-02145-6
Natural display of nuclear-encoded RNA on the cell surface and its impact on cell interaction
Abstract
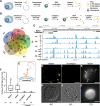
Background: Compared to proteins, glycans, and lipids, much less is known about RNAs on the cell surface. We develop a series of technologies to test for any nuclear-encoded RNAs that are stably attached to the cell surface and exposed to the extracellular space, hereafter called membrane-associated extracellular RNAs (maxRNAs).
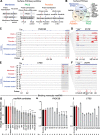
Results: We develop a technique called Surface-seq to selectively sequence maxRNAs and validate two Surface-seq identified maxRNAs by RNA fluorescence in situ hybridization. To test for cell-type specificity of maxRNA, we use antisense oligos to hybridize to single-stranded transcripts exposed on the surface of human peripheral blood mononuclear cells (PBMCs). Combining this strategy with imaging flow cytometry, single-cell RNA sequencing, and maxRNA sequencing, we identify monocytes as the major type of maxRNA+ PBMCs and prioritize 11 candidate maxRNAs for functional tests. Extracellular application of antisense oligos of FNDC3B and CTSS transcripts inhibits monocyte adhesion to vascular endothelial cells.
Conclusions: Collectively, these data highlight maxRNAs as functional components of the cell surface, suggesting an expanded role for RNA in cell-cell and cell-environment interactions.
Keywords: Cell membrane; Cell surface; Cell-environment interaction; Endothelial cells; Extracellular RNA; Monocyte; Single cell.
Conflict of interest statement
S.Z. is a founder and board member of Genemo, Inc.
Figures


References
-
- Morozkin ES, Laktionov PP, Rykova EY, Vlassov VV. Extracellular nucleic acids in cultures of long-term cultivated eukaryotic cells. Ann N Y Acad Sci. 2004;1022:244–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

