Performance and Application of 16S rRNA Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory
- PMID: 32907806
- PMCID: PMC7484979
- DOI: 10.1128/CMR.00053-19
Performance and Application of 16S rRNA Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory
Abstract
This review provides a state-of-the-art description of the performance of Sanger cycle sequencing of the 16S rRNA gene for routine identification of bacteria in the clinical microbiology laboratory. A detailed description of the technology and current methodology is outlined with a major focus on proper data analyses and interpretation of sequences. The remainder of the article is focused on a comprehensive evaluation of the application of this method for identification of bacterial pathogens based on analyses of 16S multialignment sequences. In particular, the existing limitations of similarity within 16S for genus- and species-level differentiation of clinically relevant pathogens and the lack of sequence data currently available in public databases is highlighted. A multiyear experience is described of a large regional clinical microbiology service with direct 16S broad-range PCR followed by cycle sequencing for direct detection of pathogens in appropriate clinical samples. The ability of proteomics (matrix-assisted desorption ionization-time of flight) versus 16S sequencing for bacterial identification and genotyping is compared. Finally, the potential for whole-genome analysis by next-generation sequencing (NGS) to replace 16S sequencing for routine diagnostic use is presented for several applications, including the barriers that must be overcome to fully implement newer genomic methods in clinical microbiology. A future challenge for large clinical, reference, and research laboratories, as well as for industry, will be the translation of vast amounts of accrued NGS microbial data into convenient algorithm testing schemes for various applications (i.e., microbial identification, genotyping, and metagenomics and microbiome analyses) so that clinically relevant information can be reported to physicians in a format that is understood and actionable. These challenges will not be faced by clinical microbiologists alone but by every scientist involved in a domain where natural diversity of genes and gene sequences plays a critical role in disease, health, pathogenicity, epidemiology, and other aspects of life-forms. Overcoming these challenges will require global multidisciplinary efforts across fields that do not normally interact with the clinical arena to make vast amounts of sequencing data clinically interpretable and actionable at the bedside.
Keywords: 16S rRNA; bacteria; cycle sequencing; identification.
Copyright © 2020 American Society for Microbiology.
Figures

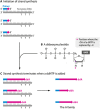

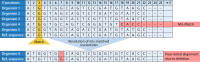
References
-
- Clinical Laboratory Standards Institute (CLSI). 2018. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; MM18-A2 approved guideline. CLSI, Wayne, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

