Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score
- PMID: 32907855
- PMCID: PMC7116472
- DOI: 10.1136/bmj.m3339
Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score
Erratum in
-
Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score.BMJ. 2020 Nov 13;371:m4334. doi: 10.1136/bmj.m4334. BMJ. 2020. PMID: 33187971 Free PMC article. No abstract available.
Abstract
Objective: To develop and validate a pragmatic risk score to predict mortality in patients admitted to hospital with coronavirus disease 2019 (covid-19).
Design: Prospective observational cohort study.
Setting: International Severe Acute Respiratory and emerging Infections Consortium (ISARIC) World Health Organization (WHO) Clinical Characterisation Protocol UK (CCP-UK) study (performed by the ISARIC Coronavirus Clinical Characterisation Consortium-ISARIC-4C) in 260 hospitals across England, Scotland, and Wales. Model training was performed on a cohort of patients recruited between 6 February and 20 May 2020, with validation conducted on a second cohort of patients recruited after model development between 21 May and 29 June 2020. PARTICIPANTS: Adults (age ≥18 years) admitted to hospital with covid-19 at least four weeks before final data extraction.
Main outcome measure: In-hospital mortality.
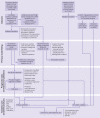
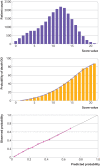
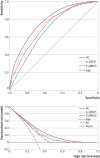
Results: 35 463 patients were included in the derivation dataset (mortality rate 32.2%) and 22 361 in the validation dataset (mortality rate 30.1%). The final 4C Mortality Score included eight variables readily available at initial hospital assessment: age, sex, number of comorbidities, respiratory rate, peripheral oxygen saturation, level of consciousness, urea level, and C reactive protein (score range 0-21 points). The 4C Score showed high discrimination for mortality (derivation cohort: area under the receiver operating characteristic curve 0.79, 95% confidence interval 0.78 to 0.79; validation cohort: 0.77, 0.76 to 0.77) with excellent calibration (validation: calibration-in-the-large=0, slope=1.0). Patients with a score of at least 15 (n=4158, 19%) had a 62% mortality (positive predictive value 62%) compared with 1% mortality for those with a score of 3 or less (n=1650, 7%; negative predictive value 99%). Discriminatory performance was higher than 15 pre-existing risk stratification scores (area under the receiver operating characteristic curve range 0.61-0.76), with scores developed in other covid-19 cohorts often performing poorly (range 0.63-0.73).
Conclusions: An easy-to-use risk stratification score has been developed and validated based on commonly available parameters at hospital presentation. The 4C Mortality Score outperformed existing scores, showed utility to directly inform clinical decision making, and can be used to stratify patients admitted to hospital with covid-19 into different management groups. The score should be further validated to determine its applicability in other populations.
Study registration: ISRCTN66726260.
© Author(s) (or their employer(s)) 2019. Re-use permitted under CC BY. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf and declare: support from the National Institute for Health Research (NIHR), the Medical Research Council (MRC), the NIHR Health Protection Research Unit (HPRU) in Emerging and Zoonotic Infections at University of Liverpool, Public Health England (PHE), Liverpool School of Tropical Medicine, University of Oxford, NIHR HPRU in Respiratory Infections at Imperial College London, Wellcome Trust, Department for International Development, the Bill and Melinda Gates Foundation, Liverpool Experimental Cancer Medicine Centre, NIHR Biomedical Research Centre at Imperial College London, EU Platform for European Preparedness Against (Re-)emerging Epidemics (PREPARE), and NIHR Clinical Research Network for the submitted work; ABD reports grants from Department of Health and Social Care (DHSC), during the conduct of the study, grants from Wellcome Trust, outside the submitted work; CAG reports grants from DHSC National Institute of Health Research (NIHR) UK, during the conduct of the study; PWH reports grants from Wellcome Trust, Department for International Development, Bill and Melinda Gates Foundation, NIHR, during the conduct of the study; JSN-V-T reports grants from DHSC, England, during the conduct of the study, and is seconded to DHSC, England; MN is supported by a Wellcome Trust investigator award and the NIHR University College London Hospitals Biomedical Research Centre (BRC); PJMO reports personal fees from consultancies and from European Respiratory Society, grants from MRC, MRC Global Challenge Research Fund, EU, NIHR BRC, MRC/GSK, Wellcome Trust, NIHR (Health Protection Research Unit (HPRU) in Respiratory Infection), and is NIHR senior investigator outside the submitted work; his role as President of the British Society for Immunology was unpaid but travel and accommodation at some meetings was provided by the Society; JKB reports grants from MRC UK; MGS reports grants from DHSC NIHR UK, grants from MRC UK, grants from HPRU in Emerging and Zoonotic Infections, University of Liverpool, during the conduct of the study, other from Integrum Scientific LLC, Greensboro, NC, USA, outside the submitted work; LCWT reports grants from HPRU in Emerging and Zoonotic Infections, University of Liverpool, during the conduct of the study, and grants from Wellcome Trust outside the submitted work; EMH, HEH, JD, RG, RP, LN, KAH, GC, LM, SH, CJ, and CG, all declare: no support from any organisation for the submitted work; no financial relationships with any organisations that might have an interest in the submitted work in the previous three years; and no other relationships or activities that could appear to have influenced the submitted work.
Figures
References
-
- Johns Hopkins Coronavirus Resource Center. COVID-19 map. 2020. https://coronavirus.jhu.edu/map.html
-
- WHO. COVID-19 situation reports. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio...
-
- Wynants L, Calster BV, Bonten MMJ, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. 2020. https://www.bmj.com/content/369/bmj.m1328 - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
- IS-HPU-1112-10117/DH_/Department of Health/United Kingdom
- RP-2016-07-012/DH_/Department of Health/United Kingdom
- MR/S032304/1/MRC_/Medical Research Council/United Kingdom
- 205228/WT_/Wellcome Trust/United Kingdom
- MR/V033441/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_19026/MRC_/Medical Research Council/United Kingdom
- MC_PC_19059/MRC_/Medical Research Council/United Kingdom
- MR/R005982/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12014/9/MRC_/Medical Research Council/United Kingdom
- DRF-2018-11-ST2-004/DH_/Department of Health/United Kingdom
- 207511/WT_/Wellcome Trust/United Kingdom
- 215091/WT_/Wellcome Trust/United Kingdom
- MC_PC_19025/MRC_/Medical Research Council/United Kingdom
- MC_PC_15001/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
