Interactions of ferritin with scavenger receptor class A members
- PMID: 32907880
- PMCID: PMC7667961
- DOI: 10.1074/jbc.RA120.014690
Interactions of ferritin with scavenger receptor class A members
Abstract
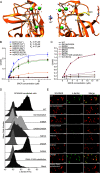
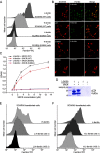
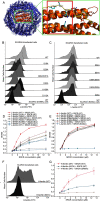
Scavenger receptors are a superfamily of membrane-bound receptors that recognize both self and nonself targets. Scavenger receptor class A (SR-A) has five known members (SCARA1 to -5 or SR-A1 to -A5), which are type II transmembrane proteins that form homotrimers on the cell surface. SR-A members recognize various ligands and are involved in multiple biological pathways. Among them, SCARA5 can function as a ferritin receptor; however, the interaction between SCARA5 and ferritin has not been fully characterized. Here, we determine the crystal structures of the C-terminal scavenger receptor cysteine-rich (SRCR) domain of both human and mouse SCARA5 at 1.7 and 2.5 Å resolution, respectively, revealing three Ca2+-binding sites on the surface. Using biochemical assays, we show that the SRCR domain of SCARA5 recognizes ferritin in a Ca2+-dependent manner, and both L- and H-ferritin can be recognized by SCARA5 through the SRCR domain. Furthermore, the potential binding region of SCARA5 on the surface of ferritin is explored by mutagenesis studies. We also examine the interactions of ferritin with other SR-A members and find that SCARA1 (SR-A1, CD204) and MARCO (SR-A2, SCARA2), which are highly expressed on macrophages, also interact with ferritin. By contrast, SCARA3 and SCARA4, the two SR-A members without the SRCR domain, have no detectable binding with ferritin. Overall, these results provide a mechanistic view regarding the interactions between the SR-A members and ferritin that may help to understand the regulation of ferritin homeostasis by scavenger receptors.
Keywords: SCARA5; SR class A; SRCR domain; X-ray crystallography; cell surface receptor; ferritin; protein structure; scavenger receptor.
© 2020 Yu et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
Recognition of lipoproteins by scavenger receptor class A members.J Biol Chem. 2021 Aug;297(2):100948. doi: 10.1016/j.jbc.2021.100948. Epub 2021 Jul 9. J Biol Chem. 2021. PMID: 34252459 Free PMC article.
-
Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells.J Biol Chem. 2006 Apr 28;281(17):11834-45. doi: 10.1074/jbc.M507599200. Epub 2006 Jan 6. J Biol Chem. 2006. PMID: 16407294
-
The scavenger receptor SCARA1 (CD204) recognizes dead cells through spectrin.J Biol Chem. 2019 Dec 6;294(49):18881-18897. doi: 10.1074/jbc.RA119.010110. Epub 2019 Oct 25. J Biol Chem. 2019. PMID: 31653705 Free PMC article.
-
The Scavenger Receptor Cysteine-Rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system.Crit Rev Immunol. 2004;24(1):1-37. doi: 10.1615/critrevimmunol.v24.i1.10. Crit Rev Immunol. 2004. PMID: 14995912 Review.
-
[The role of the class A scavenger receptors, SR-A and MARCO, in the immune system. Part 1. The structure of receptors, their ligand binding repertoires and ability to initiate intracellular signaling].Postepy Hig Med Dosw (Online). 2012 Feb 29;66:104-19. doi: 10.5604/17322693.984079. Postepy Hig Med Dosw (Online). 2012. PMID: 22470185 Review. Polish.
Cited by
-
Therapeutic potential of induced iron depletion using iron chelators in Covid-19.Saudi J Biol Sci. 2022 Apr;29(4):1947-1956. doi: 10.1016/j.sjbs.2021.11.061. Epub 2021 Dec 13. Saudi J Biol Sci. 2022. PMID: 34924800 Free PMC article. Review.
-
Danger-Sensing/Patten Recognition Receptors and Neuroinflammation in Alzheimer's Disease.Int J Mol Sci. 2020 Nov 27;21(23):9036. doi: 10.3390/ijms21239036. Int J Mol Sci. 2020. PMID: 33261147 Free PMC article. Review.
-
Ferritin in Acute Myeloid Leukemia: Not Only a Marker of Inflammation and Iron Overload, but Also a Regulator of Cellular Iron Metabolism, Signaling and Communication.Int J Mol Sci. 2025 Jun 15;26(12):5744. doi: 10.3390/ijms26125744. Int J Mol Sci. 2025. PMID: 40565207 Free PMC article. Review.
-
The Role of SCARA5 as a Potential Biomarker in Squamous Cell Carcinoma of the Lung.Int J Mol Sci. 2024 Jul 4;25(13):7355. doi: 10.3390/ijms25137355. Int J Mol Sci. 2024. PMID: 39000462 Free PMC article.
-
An interpretable machine learning-based cerebrospinal fluid proteomics clock for predicting age reveals novel insights into brain aging.Aging Cell. 2024 Sep;23(9):e14230. doi: 10.1111/acel.14230. Epub 2024 Jun 24. Aging Cell. 2024. PMID: 38923730 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

