Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids
- PMID: 32907944
- PMCID: PMC7519316
- DOI: 10.1073/pnas.2010834117
Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids
Abstract
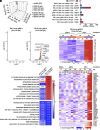
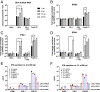
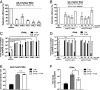
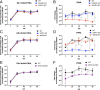
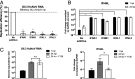
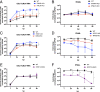
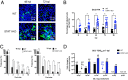
Human noroviruses (HuNoVs) are the leading cause of viral gastroenteritis worldwide; yet currently, no vaccines or FDA-approved antiviral drugs are available to counter these pathogens. To understand HuNoV biology and the epithelial response to infection, we performed transcriptomic analyses, RT-qPCR, CRISPR-Cas9 modification of human intestinal enteroid (HIE) cultures, and functional studies with two virus strains (a pandemic GII.4 and a bile acid-dependent GII.3 strain). We identified a predominant type III interferon (IFN)-mediated innate response to HuNoV infection. Replication of both strains is sensitive to exogenous addition of IFNs, suggesting the potential of IFNs as therapeutics. To obtain insight into IFN pathway genes that play a role in the antiviral response to HuNoVs, we developed knockout (KO) HIE lines for IFN alpha and lambda receptors and the signaling molecules, MAVS, STAT1, and STAT2 An unexpected differential response of enhanced replication and virus spread was observed for GII.3, but not the globally dominant GII.4 HuNoV in STAT1-knockout HIEs compared to parental HIEs. These results indicate cellular IFN responses restrict GII.3 but not GII.4 replication. The strain-specific sensitivities of innate responses against HuNoV replication provide one explanation for why GII.4 infections are more widespread and highlight strain specificity as an important factor in HuNoV biology. Genetically modified HIEs for innate immune genes are useful tools for studying immune responses to viral or microbial pathogens.
Keywords: CRISPR-Cas9; RNA-Seq; enteroids/organoids; human norovirus; interferon.
Conflict of interest statement
Competing interest statement: M.K.E. is named as an inventor on patents related to cloning and cultivation of the Norwalk virus genome and is a consultant to and received research funding from Takeda Vaccines, Inc. R.L.A. has received research funding from Takeda Vaccines, Inc.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

