Dolosigranulum pigrum Cooperation and Competition in Human Nasal Microbiota
- PMID: 32907957
- PMCID: PMC7485692
- DOI: 10.1128/mSphere.00852-20
Dolosigranulum pigrum Cooperation and Competition in Human Nasal Microbiota
Abstract
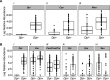
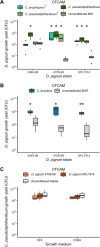
Multiple epidemiological studies identify Dolosigranulum pigrum as a candidate beneficial bacterium based on its positive association with health, including negative associations with nasal/nasopharyngeal colonization by the pathogenic species Staphylococcus aureus and Streptococcus pneumoniae Using a multipronged approach to gain new insights into D. pigrum function, we observed phenotypic interactions and predictions of genomic capacity that support the idea of a role for microbe-microbe interactions involving D. pigrum in shaping the composition of human nasal microbiota. We identified in vivo community-level and in vitro phenotypic cooperation by specific nasal Corynebacterium species. Also, D. pigrum inhibited S. aureus growth in vitro, whereas robust inhibition of S. pneumoniae required both D. pigrum and a nasal Corynebacterium together. D. pigrum l-lactic acid production was insufficient to account for these inhibitions. Genomic analysis of 11 strains revealed that D. pigrum has a small genome (average 1.86 Mb) and multiple predicted auxotrophies consistent with D. pigrum relying on its human host and on cocolonizing bacteria for key nutrients. Further, the accessory genome of D. pigrum harbored a diverse repertoire of biosynthetic gene clusters, some of which may have a role in microbe-microbe interactions. These new insights into D. pigrum's functions advance the field from compositional analysis to genomic and phenotypic experimentation on a potentially beneficial bacterial resident of the human upper respiratory tract and lay the foundation for future animal and clinical experiments.IMPORTANCEStaphylococcus aureus and Streptococcus pneumoniae infections cause significant morbidity and mortality in humans. For both, nasal colonization is a risk factor for infection. Studies of nasal microbiota identify Dolosigranulum pigrum as a benign bacterium present when adults are free of S. aureus or when children are free of S. pneumoniae Here, we validated these in vivo associations with functional assays. We found that D. pigrum inhibited S. aureusin vitro and, together with a specific nasal Corynebacterium species, also inhibited S. pneumoniae Furthermore, genomic analysis of D. pigrum indicated that it must obtain key nutrients from other nasal bacteria or from humans. These phenotypic interactions support the idea of a role for microbe-microbe interactions in shaping the composition of human nasal microbiota and implicate D. pigrum as a mutualist of humans. These findings support the feasibility of future development of microbe-targeted interventions to reshape nasal microbiota composition to exclude S. aureus and/or S. pneumoniae.
Keywords: Corynebacterium; Dolosigranulum pigrum; Staphylococcus aureus; Streptococcus pneumoniae; comparative genomics; interspecies interactions; microbe-microbe interactions; microbiota; nasal; upper respiratory tract.
Copyright © 2020 Brugger et al.
Figures



References
-
- Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705. doi: 10.1016/S0140-6736(04)16897-9. - DOI - PubMed
-
- Young BC, Wu CH, Gordon NC, Cole K, Price JR, Liu E, Sheppard AE, Perera S, Charlesworth J, Golubchik T, Iqbal Z, Bowden R, Massey RC, Paul J, Crook DW, Peto TE, Walker AS, Llewelyn MJ, Wyllie DH, Wilson DJ. 2017. Severe infections emerge from commensal bacteria by adaptive evolution. Elife 6:e30637. doi: 10.7554/eLife.30637. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

