Antigenic Restimulation of Virus-Specific Memory CD8+ T Cells Requires Days of Lytic Protein Accumulation for Maximal Cytotoxic Capacity
- PMID: 32907983
- PMCID: PMC7654275
- DOI: 10.1128/JVI.01595-20
Antigenic Restimulation of Virus-Specific Memory CD8+ T Cells Requires Days of Lytic Protein Accumulation for Maximal Cytotoxic Capacity
Abstract
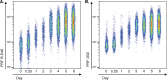
In various infections or vaccinations of mice or humans, reports of the persistence and the requirements for restimulation of the cytotoxic mediators granzyme B (GrB) and perforin (PRF) in CD8+ T cells have yielded disparate results. In this study, we examined the kinetics of PRF and GrB mRNA and protein expression after stimulation and associated changes in cytotoxic capacity in virus-specific memory cells in detail. In patients with controlled HIV or cleared respiratory syncytial virus (RSV) or influenza virus infections, all virus-specific CD8+ T cells expressed low PRF levels without restimulation. Following stimulation, they displayed similarly delayed kinetics for lytic protein expression, with significant increases occurring by days 1 to 3 before peaking on days 4 to 6. These increases were strongly correlated with, but were not dependent upon, proliferation. Incremental changes in PRF and GrB percent expression and mean fluorescence intensity (MFI) were highly correlated with increases in HIV-specific cytotoxicity. mRNA levels in HIV-specific CD8+ T-cells exhibited delayed kinetics after stimulation as with protein expression, peaking on day 5. In contrast to GrB, PRF mRNA transcripts were little changed over 5 days of stimulation (94-fold versus 2.8-fold, respectively), consistent with posttranscriptional regulation. Changes in expression of some microRNAs, including miR-17, miR-150, and miR-155, suggested that microRNAs might play a significant role in regulation of PRF expression. Therefore, under conditions of extremely low or absent antigen levels, memory virus-specific CD8+ T cells require prolonged stimulation over days to achieve maximal lytic protein expression and cytotoxic capacity.IMPORTANCE Antigen-specific CD8+ T cells play a major role in controlling most virus infections, primarily by perforin (PRF)- and granzyme B (GrB)-mediated apoptosis. There is considerable controversy regarding whether PRF is constitutively expressed, rapidly increased similarly to a cytokine, or delayed in its expression with more prolonged stimulation in virus-specific memory CD8+ T cells. In this study, the degree of cytotoxic capacity of virus-specific memory CD8+ T cells was directly proportional to the content of lytic molecules, which required antigenic stimulation over several days for maximal levels. This appeared to be modulated by increases in GrB transcription and microRNA-mediated posttranscriptional regulation of PRF expression. Clarifying the requirements for maximal cytotoxic capacity is critical to understanding how viral clearance might be mediated by memory cells and what functions should be induced by vaccines and immunotherapies.
Keywords: CD8+ T cells; kinetics; perforin; virus-specific immunity.
Copyright © 2020 American Society for Microbiology.
Figures






References
-
- Wang Z, Wan Y, Qiu C, Quinones-Parra S, Zhu Z, Loh L, Tian D, Ren Y, Hu Y, Zhang X, Thomas PG, Inouye M, Doherty PC, Kedzierska K, Xu J. 2015. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8(+) T cells. Nat Commun 6:6833. doi: 10.1038/ncomms7833. - DOI - PMC - PubMed
-
- Jozwik A, Habibi MS, Paras A, Zhu J, Guvenel A, Dhariwal J, Almond M, Wong EH, Sykes A, Maybeno M, Del Rosario J, Trujillo-Torralbo MB, Mallia P, Sidney J, Peters B, Kon OM, Sette A, Johnston SL, Openshaw PJ, Chiu C. 2015. RSV-specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat Commun 6:10224. doi: 10.1038/ncomms10224. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

