A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts
- PMID: 32908151
- PMCID: PMC7481185
- DOI: 10.1038/s41467-020-18234-w
A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts
Abstract
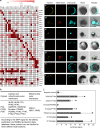
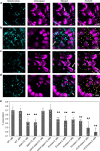
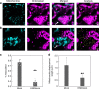
Glycolysis is one of the primordial pathways of metabolism, playing a pivotal role in energy metabolism and biosynthesis. Glycolytic enzymes are known to form transient multi-enzyme assemblies. Here we examine the wider protein-protein interactions of plant glycolytic enzymes and reveal a moonlighting role for specific glycolytic enzymes in mediating the co-localization of mitochondria and chloroplasts. Knockout mutation of phosphoglycerate mutase or enolase resulted in a significantly reduced association of the two organelles. We provide evidence that phosphoglycerate mutase and enolase form a substrate-channelling metabolon which is part of a larger complex of proteins including pyruvate kinase. These results alongside a range of genetic complementation experiments are discussed in the context of our current understanding of chloroplast-mitochondrial interactions within photosynthetic eukaryotes.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Kohler R. The background to Eduard Buchner’s discovery of cell-free fermentation. J. Hist. Biol. 1971;4:35–61. - PubMed
-
- Plaxton WC. The organization and regulation of plant glycolysis. Annu. Rev. Plant Biol. 1996;47:185–214. - PubMed
-
- Araiza Olivera D, et al. A glycolytic metabolon in Saccharomyces cerevisiae is stabilized by F-actin. FEBS J. 2013;280:3887–3905. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

