TRIM37 controls cancer-specific vulnerability to PLK4 inhibition
- PMID: 32908304
- PMCID: PMC7501188
- DOI: 10.1038/s41586-020-2710-1
TRIM37 controls cancer-specific vulnerability to PLK4 inhibition
Abstract
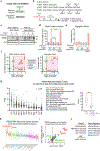
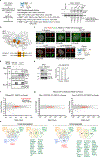
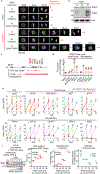
Centrosomes catalyse the formation of microtubules needed to assemble the mitotic spindle apparatus1. Centrosomes themselves duplicate once per cell cycle, in a process that is controlled by the serine/threonine protein kinase PLK4 (refs. 2,3). When PLK4 is chemically inhibited, cell division proceeds without centrosome duplication, generating centrosome-less cells that exhibit delayed, acentrosomal spindle assembly4. Whether PLK4 inhibitors can be leveraged as a treatment for cancer is not yet clear. Here we show that acentrosomal spindle assembly following PLK4 inhibition depends on levels of the centrosomal ubiquitin ligase TRIM37. Low TRIM37 levels accelerate acentrosomal spindle assembly and improve proliferation following PLK4 inhibition, whereas high TRIM37 levels inhibit acentrosomal spindle assembly, leading to mitotic failure and cessation of proliferation. The Chr17q region containing the TRIM37 gene is frequently amplified in neuroblastoma and in breast cancer5-8, rendering these cancer types highly sensitive to PLK4 inhibition. We find that inactivating TRIM37 improves acentrosomal mitosis because TRIM37 prevents PLK4 from self-assembling into centrosome-independent condensates that serve as ectopic microtubule-organizing centres. By contrast, elevated TRIM37 expression inhibits acentrosomal spindle assembly through a distinct mechanism that involves degradation of the centrosomal component CEP192. Thus, TRIM37 is an essential determinant of mitotic vulnerability to PLK4 inhibition. Linkage of TRIM37 to prevalent cancer-associated genomic changes-including 17q gain in neuroblastoma and 17q23 amplification in breast cancer-may offer an opportunity to use PLK4 inhibition to trigger selective mitotic failure and provide new avenues to treatments for these cancers.
Conflict of interest statement
COMPETING INTERESTS
K.O., A.D., A.K.S., R.L.D., and F.M. are inventors on a pending patent application from the Ludwig Institute for Cancer Research Ltd, application number PCT/US2018/026720. “TRIM37 levels predict sensitivity to PLK4 inhibition in cancer cells”.
Figures








Comment in
-
Selectively triggering mitotic failure.Nat Rev Drug Discov. 2020 Nov;19(11):755. doi: 10.1038/d41573-020-00165-2. Nat Rev Drug Discov. 2020. PMID: 32943764 No abstract available.
Similar articles
-
Targeting TRIM37-driven centrosome dysfunction in 17q23-amplified breast cancer.Nature. 2020 Sep;585(7825):447-452. doi: 10.1038/s41586-020-2690-1. Epub 2020 Sep 9. Nature. 2020. PMID: 32908313 Free PMC article.
-
Global cellular response to chemical perturbation of PLK4 activity and abnormal centrosome number.Elife. 2022 Jun 27;11:e73944. doi: 10.7554/eLife.73944. Elife. 2022. PMID: 35758262 Free PMC article.
-
TRIM37: a critical orchestrator of centrosome function.Cell Cycle. 2021 Dec;20(23):2443-2451. doi: 10.1080/15384101.2021.1988289. Epub 2021 Oct 21. Cell Cycle. 2021. PMID: 34672905 Free PMC article. Review.
-
TRIM37 prevents formation of condensate-organized ectopic spindle poles to ensure mitotic fidelity.J Cell Biol. 2021 Jul 5;220(7):e202010180. doi: 10.1083/jcb.202010180. Epub 2021 May 13. J Cell Biol. 2021. PMID: 33983387 Free PMC article.
-
PLK4: Master Regulator of Centriole Duplication and Its Therapeutic Potential.Cytoskeleton (Hoboken). 2025 Apr 21. doi: 10.1002/cm.22031. Online ahead of print. Cytoskeleton (Hoboken). 2025. PMID: 40257113 Review.
Cited by
-
TFEB- and TFE3-dependent autophagy activation supports cancer proliferation in the absence of centrosomes.Autophagy. 2022 Dec;18(12):2830-2850. doi: 10.1080/15548627.2022.2051880. Epub 2022 Mar 22. Autophagy. 2022. PMID: 35316161 Free PMC article.
-
Centriolar subdistal appendages promote double-strand break repair through homologous recombination.EMBO Rep. 2023 Oct 9;24(10):e56724. doi: 10.15252/embr.202256724. Epub 2023 Sep 4. EMBO Rep. 2023. PMID: 37664992 Free PMC article.
-
Chromosomal Instability and Clonal Heterogeneity in Breast Cancer: From Mechanisms to Clinical Applications.Cancers (Basel). 2025 Apr 4;17(7):1222. doi: 10.3390/cancers17071222. Cancers (Basel). 2025. PMID: 40227811 Free PMC article. Review.
-
Biomolecular phase separation in tumorigenesis: from aberrant condensates to therapeutic vulnerabilities.Mol Cancer. 2025 Aug 23;24(1):220. doi: 10.1186/s12943-025-02428-1. Mol Cancer. 2025. PMID: 40849651 Free PMC article. Review.
-
Non-proteolytic ubiquitylation in cellular signaling and human disease.Commun Biol. 2022 Feb 8;5(1):114. doi: 10.1038/s42003-022-03060-1. Commun Biol. 2022. PMID: 35136173 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

