Targeting TRIM37-driven centrosome dysfunction in 17q23-amplified breast cancer
- PMID: 32908313
- PMCID: PMC7597367
- DOI: 10.1038/s41586-020-2690-1
Targeting TRIM37-driven centrosome dysfunction in 17q23-amplified breast cancer
Abstract
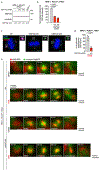
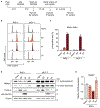
Genomic instability is a hallmark of cancer, and has a central role in the initiation and development of breast cancer1,2. The success of poly-ADP ribose polymerase inhibitors in the treatment of breast cancers that are deficient in homologous recombination exemplifies the utility of synthetically lethal genetic interactions in the treatment of breast cancers that are driven by genomic instability3. Given that defects in homologous recombination are present in only a subset of breast cancers, there is a need to identify additional driver mechanisms for genomic instability and targeted strategies to exploit these defects in the treatment of cancer. Here we show that centrosome depletion induces synthetic lethality in cancer cells that contain the 17q23 amplicon, a recurrent copy number aberration that defines about 9% of all primary breast cancer tumours and is associated with high levels of genomic instability4-6. Specifically, inhibition of polo-like kinase 4 (PLK4) using small molecules leads to centrosome depletion, which triggers mitotic catastrophe in cells that exhibit amplicon-directed overexpression of TRIM37. To explain this effect, we identify TRIM37 as a negative regulator of centrosomal pericentriolar material. In 17q23-amplified cells that lack centrosomes, increased levels of TRIM37 block the formation of foci that comprise pericentriolar material-these foci are structures with a microtubule-nucleating capacity that are required for successful cell division in the absence of centrosomes. Finally, we find that the overexpression of TRIM37 causes genomic instability by delaying centrosome maturation and separation at mitotic entry, and thereby increases the frequency of mitotic errors. Collectively, these findings highlight TRIM37-dependent genomic instability as a putative driver event in 17q23-amplified breast cancer and provide a rationale for the use of centrosome-targeting therapeutic agents in treating these cancers.
Figures












Comment in
-
Selectively triggering mitotic failure.Nat Rev Drug Discov. 2020 Nov;19(11):755. doi: 10.1038/d41573-020-00165-2. Nat Rev Drug Discov. 2020. PMID: 32943764 No abstract available.
References
-
- Curtis C et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346, doi: 10.1038/nature10983 https://www.nature.com/articles/nature10983#supplementary-information (2012). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

