Concordance of Genomic Alterations between Circulating Tumor DNA and Matched Tumor Tissue in Chinese Patients with Breast Cancer
- PMID: 32908507
- PMCID: PMC7474381
- DOI: 10.1155/2020/4259293
Concordance of Genomic Alterations between Circulating Tumor DNA and Matched Tumor Tissue in Chinese Patients with Breast Cancer
Abstract
Purpose: Circulating tumor DNA (ctDNA) served as a noninvasive method with less side effects using peripheral blood. Given the studies on concordance rate between liquid and solid biopsies in Chinese breast cancer (BC) patients were limited, we sought to examine the concordance rate of different kinds of genomic alterations between paired tissue biopsies and ctDNA samples in Chinese BC cohorts.
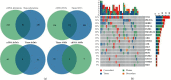
Materials and methods: In this study, we analyzed the genomic alteration profiles of 81 solid BC samples and 41 liquid BC samples. The concordance across 136 genes was evaluated.
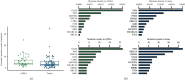
Results: The median mutation counts per sample in 41 ctDNA samples was higher than the median in 81 tissue samples (p=0.0254; Wilcoxon rank sum test). For mutation at the protein-coding level, 39.0% (16/41) samples had at least one concordant mutation in two biopsies. 20.0% tissue-derived mutations could be detected via ctDNA-based sequencing, whereas 11.7% ctDNA-derived mutations could be found in paired tissues. At gene amplification level, the overall concordant rate was 68.3% (28/41). The concordant rate at gene level for each patient ranged from 83.8% (114/136) to 99.3% (135/136). And, the mean level of variant allele frequency (VAF) for concordant mutations in ctDNA was statistically higher than that for the discordant ones (p < 0.001; Wilcoxon rank sum test). Across five representative genes, the overall sensitivity and specificity were 49.0% and 85.9%, respectively.
Conclusion: Our results indicated that ctDNA could provide complementary information on genetic characterizations in detecting single nucleotide variants (SNVs) and insertions and deletions (InDels).
Copyright © 2020 Bing Xu et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Figures

Similar articles
-
Concordance of Genomic Alterations by Next-Generation Sequencing in Tumor Tissue versus Circulating Tumor DNA in Breast Cancer.Mol Cancer Ther. 2017 Jul;16(7):1412-1420. doi: 10.1158/1535-7163.MCT-17-0061. Epub 2017 Apr 26. Mol Cancer Ther. 2017. PMID: 28446639
-
High concordance of actionable genomic alterations identified between circulating tumor DNA-based and tissue-based next-generation sequencing testing in advanced non-small cell lung cancer: The Korean Lung Liquid Versus Invasive Biopsy Program.Cancer. 2021 Aug 15;127(16):3019-3028. doi: 10.1002/cncr.33571. Epub 2021 Apr 7. Cancer. 2021. PMID: 33826761
-
Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer.J Natl Cancer Inst. 2017 Dec 1;109(12):djx118. doi: 10.1093/jnci/djx118. J Natl Cancer Inst. 2017. PMID: 29206995 Free PMC article.
-
Guardant360 Circulating Tumor DNA Assay Is Concordant with FoundationOne Next-Generation Sequencing in Detecting Actionable Driver Mutations in Anti-EGFR Naive Metastatic Colorectal Cancer.Oncologist. 2020 Mar;25(3):235-243. doi: 10.1634/theoncologist.2019-0441. Epub 2019 Nov 19. Oncologist. 2020. PMID: 32162812 Free PMC article.
-
Genomic profiling of cell-free circulating tumor DNA in patients with colorectal cancer and its fidelity to the genomics of the tumor biopsy.J Gastrointest Oncol. 2019 Oct;10(5):831-840. doi: 10.21037/jgo.2019.05.05. J Gastrointest Oncol. 2019. PMID: 31602320 Free PMC article.
Cited by
-
Concordance between whole exome sequencing of circulating tumor DNA and tumor tissue.PLoS One. 2023 Oct 25;18(10):e0292879. doi: 10.1371/journal.pone.0292879. eCollection 2023. PLoS One. 2023. PMID: 37878600 Free PMC article.
-
Analytical and clinical validation of a custom 15-gene next-generation sequencing panel for the evaluation of circulating tumor DNA mutations in patients with advanced non-small-cell lung cancer.PLoS One. 2022 Oct 18;17(10):e0276161. doi: 10.1371/journal.pone.0276161. eCollection 2022. PLoS One. 2022. PMID: 36256645 Free PMC article.
-
Mutation analysis of circulating tumor DNA and paired ascites and tumor tissues in ovarian cancer.Exp Ther Med. 2022 Jun 30;24(3):542. doi: 10.3892/etm.2022.11479. eCollection 2022 Sep. Exp Ther Med. 2022. PMID: 35978934 Free PMC article.
-
Longitudinal Shifts of Solid Tumor and Liquid Biopsy Sequencing Concordance in Metastatic Breast Cancer.JCO Precis Oncol. 2022 Jun 6;6(1):e2100321. doi: 10.1200/PO.21.00321. eCollection 2022 Jun. JCO Precis Oncol. 2022. PMID: 35721584 Free PMC article.
-
How to Predict Metastasis in Luminal Breast Cancer? Current Solutions and Future Prospects.Int J Mol Sci. 2020 Nov 9;21(21):8415. doi: 10.3390/ijms21218415. Int J Mol Sci. 2020. PMID: 33182512 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

