Genomic and Pathogenic Characteristics of Virulent Newcastle Disease Virus Isolated from Chicken in Live Bird Markets and Backyard Flocks in Kenya
- PMID: 32908524
- PMCID: PMC7450340
- DOI: 10.1155/2020/4705768
Genomic and Pathogenic Characteristics of Virulent Newcastle Disease Virus Isolated from Chicken in Live Bird Markets and Backyard Flocks in Kenya
Abstract
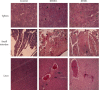
Newcastle disease (ND) causes significant economic losses in the poultry industry in developing countries. In Kenya, despite rampant annual ND outbreaks, implementation of control strategies is hampered by a lack of adequate knowledge on the circulating and outbreak causing-NDV strains. This study reports the first complete genome sequences of NDV from backyard chicken in Kenya. The results showed that all three isolates are virulent, as assessed by the mean death time (MDT) and intracerebral pathogenicity index (ICPI) in specific antibody negative (SAN) embryonated eggs and 10-day-old chickens, respectively. Also, the polybasic amino acid sequence at the fusion-protein cleavage site had the motif 112RRQKRFV118. Histopathological findings in four-week-old SPF chicken challenged with the NDV isolates KE001, KE0811, and KE0698 showed multiple organ involvement at five days after infection with severe effects seen in lymphoid tissues and blood vessels. Analysis of genome sequences obtained from the three isolates showed that they were 15192 base pair (bp) in length and had genomic features consistent with other NDV strains, the functional sites within the coding sequence being highly conserved in the sequence of the three isolates. Amino acid residues and substitutions in the structural proteins of the three isolates were similar to the newly isolated Tanzanian NDV strain (Mbeya/MT15). A similarity matrix showed a high similarity of the isolates to NDV strains of class II genotype V (89-90%) and subgenotype Vd (95-97%). Phylogenetic analysis confirmed that the three isolates are closely related to NDV genotype V strains but form a distinct cluster together with NDV strains from the East African countries of Uganda and Tanzania to form the newly characterized subgenotype Vd. Our study provides the first description of the genomic and pathological characteristics of NDV of subgenotype Vd and lays a baseline in understanding the evolutionary dynamics of NDV and, in particular, Genotype V. This information will be useful in the development of specific markers for detection of viruses of genotype V and generation of genotype matched vaccines.
Copyright © 2020 Irene N. Ogali et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Figures



References
-
- OIE. Newcastle disease: general infectious disease sheets. 2015. https://www.oie.int/fileadmin/Home/eng/Media_Center/docs/pdf/Disease_car....
-
- Ashraf A., Shah M. S. Newcastle disease: present status and future challenges for developing countries. African Journal of Microbiology Research. 2014;8(5):411–416. doi: 10.5897/ajmr2013.6540. - DOI
LinkOut - more resources
Full Text Sources

