Bioinformatics Analysis of Key Genes and circRNA-miRNA-mRNA Regulatory Network in Gastric Cancer
- PMID: 32908877
- PMCID: PMC7463386
- DOI: 10.1155/2020/2862701
Bioinformatics Analysis of Key Genes and circRNA-miRNA-mRNA Regulatory Network in Gastric Cancer
Abstract
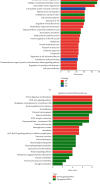
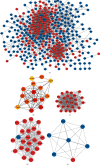
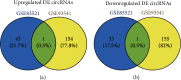
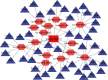
Gastric cancer (GC) is one of the most common malignancies in the world, with morbidity and mortality ranking second among all cancers. Accumulating evidences indicate that circular RNAs (circRNAs) are closely correlated with tumorigenesis. However, the mechanisms of circRNAs still remain unclear. This study is aimed at determining hub genes and circRNAs and analyzing their potential biological functions in GC. Expression profiles of mRNAs and circRNAs were downloaded from the Gene Expression Omnibus (GEO) data sets of GC and paracancer tissues. Differentially expressed genes (DEGs) and differentially expressed circRNAs (DE-circRNAs) were identified. The target miRNAs of DE-circRNAs and the bidirectional interaction between target miRNAs and DEGs were predicted. Functional analysis was performed, and the protein-protein interaction (PPI) network and the circRNA-miRNA-mRNA network were established. A total of 456 DEGs and 2 DE-circRNAs were identified with 3 mRNA expression profiles and 2 circRNA expression profiles. GO analysis indicated that DEGs were mainly enriched in extracellular matrix and cell adhesion, and KEGG confirmed that DEGs were mainly associated with focal adhesion, the PI3K-Akt signaling pathway, extracellular matrix- (ECM)- receptor interaction, and gastric acid secretion. 15 hub DEGs (BGN, COL1A1, COL1A2, FBN1, FN1, SPARC, SPP1, TIMP1, UBE2C, CCNB1, CD44, CXCL8, COL3A1, COL5A2, and THBS1) were identified from the PPI network. Furthermore, the survival analysis indicate that GC patients with a high expression of the following 9 hub DEGs, namely, BGN, COL1A1, COL1A2, FBN1, FN1, SPARC, SPP1, TIMP1, and UBE2C, had significantly worse overall survival. The circRNA-miRNA-mRNA network was constructed based on 1 circRNA, 15 miRNAs, and 45 DEGs. In addition, the 45 DEGs included 5 hub DEGs. These results suggested that hub DEGs and circRNAs could be implicated in the pathogenesis and development of GC. Our findings provide novel evidence on the circRNA-miRNA-mRNA network and lay the foundation for future research of circRNAs in GC.
Copyright © 2020 Yiting Tian et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures


References
-
- Liu W., Ouyang S., Zhou Z., et al. Identification of genes associated with cancer progression and prognosis in lung adenocarcinoma: analyses based on microarray from Oncomine and The Cancer Genome Atlas databases. Molecular genetics & genomic medicine. 2019;7(2, article e00528) doi: 10.1002/mgg3.528. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

