Disease-Associated Circular RNAs: From Biology to Computational Identification
- PMID: 32908906
- PMCID: PMC7450300
- DOI: 10.1155/2020/6798590
Disease-Associated Circular RNAs: From Biology to Computational Identification
Abstract
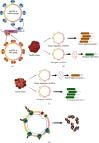
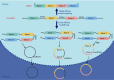
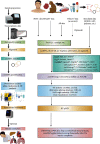
Circular RNAs (circRNAs) are endogenous RNAs with a covalently closed continuous loop, generated through various backsplicing events of pre-mRNA. An accumulating number of studies have shown that circRNAs are potential biomarkers for major human diseases such as cancer and Alzheimer's disease. Thus, identification and prediction of human disease-associated circRNAs are of significant importance. To this end, a computational analysis-assisted strategy is indispensable to detect, verify, and quantify circRNAs for downstream applications. In this review, we briefly introduce the biology of circRNAs, including the biogenesis, characteristics, and biological functions. In addition, we outline about 30 recent bioinformatic analysis tools that are publicly available for circRNA study. Principles for applying these computational strategies and considerations will be briefly discussed. Lastly, we give a complete survey on more than 20 key computational databases that are frequently used. To our knowledge, this is the most complete and updated summary on publicly available circRNA resources. In conclusion, this review summarizes key aspects of circRNA biology and outlines key computational strategies that will facilitate the genome-wide identification and prediction of circRNAs.
Copyright © 2020 Min Tang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
Similar articles
-
Insights into circular RNAs: their biogenesis, detection, and emerging role in cardiovascular disease.RNA Biol. 2021 Dec;18(12):2055-2072. doi: 10.1080/15476286.2021.1891393. Epub 2021 Mar 28. RNA Biol. 2021. PMID: 33779499 Free PMC article. Review.
-
Circular RNAs and complex diseases: from experimental results to computational models.Brief Bioinform. 2021 Nov 5;22(6):bbab286. doi: 10.1093/bib/bbab286. Brief Bioinform. 2021. PMID: 34329377 Free PMC article. Review.
-
Profiling of differentially expressed circular RNAs in peripheral blood mononuclear cells from Alzheimer's disease patients.Metab Brain Dis. 2020 Jan;35(1):201-213. doi: 10.1007/s11011-019-00497-y. Epub 2019 Dec 13. Metab Brain Dis. 2020. PMID: 31834549
-
circRNAprofiler: an R-based computational framework for the downstream analysis of circular RNAs.BMC Bioinformatics. 2020 Apr 29;21(1):164. doi: 10.1186/s12859-020-3500-3. BMC Bioinformatics. 2020. PMID: 32349660 Free PMC article.
-
Computational Prediction of Human Disease- Associated circRNAs Based on Manifold Regularization Learning Framework.IEEE J Biomed Health Inform. 2019 Nov;23(6):2661-2669. doi: 10.1109/JBHI.2019.2891779. Epub 2019 Jan 9. IEEE J Biomed Health Inform. 2019. PMID: 30629521
Cited by
-
Role of circRNA-miRNA-mRNA interaction network in diabetes and its associated complications.Mol Ther Nucleic Acids. 2021 Nov 10;26:1291-1302. doi: 10.1016/j.omtn.2021.11.007. eCollection 2021 Dec 3. Mol Ther Nucleic Acids. 2021. PMID: 34853728 Free PMC article. Review.
-
Comprehensive circular RNA profiling in various sheep tissues.Sci Rep. 2024 Oct 31;14(1):26238. doi: 10.1038/s41598-024-76940-7. Sci Rep. 2024. PMID: 39482374 Free PMC article.
-
CircAMOTL1 RNA and AMOTL1 Protein: Complex Functions of AMOTL1 Gene Products.Int J Mol Sci. 2023 Jan 20;24(3):2103. doi: 10.3390/ijms24032103. Int J Mol Sci. 2023. PMID: 36768425 Free PMC article. Review.
-
Comprehensive Analysis of RNA Expression Profile Identifies Hub miRNA-circRNA Interaction Networks in the Hypoxic Ischemic Encephalopathy.Comput Math Methods Med. 2021 Sep 21;2021:6015473. doi: 10.1155/2021/6015473. eCollection 2021. Comput Math Methods Med. 2021. PMID: 34603484 Free PMC article.
-
Hsa_circ_0001359 in Serum Exosomes: A Promising Marker to Predict Bronchopulmonary Dysplasia in Premature Infants.J Inflamm Res. 2024 Jul 25;17:5025-5037. doi: 10.2147/JIR.S463330. eCollection 2024. J Inflamm Res. 2024. PMID: 39081873 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

