The Stress-Like Cancer Cell State Is a Consistent Component of Tumorigenesis
- PMID: 32910905
- PMCID: PMC8027961
- DOI: 10.1016/j.cels.2020.08.018
The Stress-Like Cancer Cell State Is a Consistent Component of Tumorigenesis
Abstract
Transcriptional profiling of tumors has revealed a stress-like state among the cancer cells with the concerted expression of genes such as fos, jun, and heat-shock proteins, though this has been controversial given possible dissociation-effects associated with single-cell RNA sequencing. Here, we validate the existence of this state using a combination of zebrafish melanoma modeling, spatial transcriptomics, and human samples. We found that the stress-like subpopulation of cancer cells is present from the early stages of tumorigenesis. Comparing with previously reported single-cell RNA sequencing datasets from diverse cancer types, including triple-negative breast cancer, oligodendroglioma, and pancreatic adenocarcinoma, indicated the conservation of this state during tumorigenesis. We also provide evidence that this state has higher tumor-seeding capabilities and that its induction leads to increased growth under both MEK and BRAF inhibitors. Collectively, our study supports the stress-like cells as a cancer cell state expressing a coherent set of genes and exhibiting drug-resistance properties.
Keywords: cancer cell states; drug-resistant states; melanoma; single-cell RNA-seq; spatial transcriptomics; stress-like.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests R.M.W. is a paid consultant to N-of-One, Inc., a subsidiary of QIAGEN. None of the work described in this manuscript is related to this work. He serves on the Scientific Advisory Board of Consano, a non-profit crowdfunding company and receives no compensation for this work.
Figures

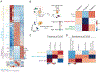
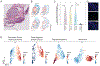

References
-
- Barkley D, and Yanai I (2019). Plasticity and Clonality of Cancer Cell States. Trends Cancer Res. 5, 655–656. - PubMed
-
- Bianchi M, Giacomini E, Crinelli R, Radici L, Carloni E, and Magnani M (2015). Dynamic transcription of ubiquitin genes under basal and stressful conditions and new insights into the multiple UBC transcript variants. Gene 573, 100–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

