Polyploidy of semi-cloned embryos generated from parthenogenetic haploid embryonic stem cells
- PMID: 32911495
- PMCID: PMC7482839
- DOI: 10.1371/journal.pone.0233072
Polyploidy of semi-cloned embryos generated from parthenogenetic haploid embryonic stem cells
Abstract
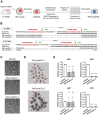
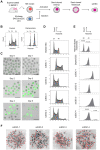
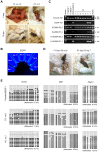
In mammals, the fusion of two gametes, an oocyte and a spermatozoon, during fertilization forms a totipotent zygote. There has been no reported case of adult mammal development by natural parthenogenesis, in which embryos develop from unfertilized oocytes. The genome and epigenetic information of haploid gametes are crucial for mammalian development. Haploid embryonic stem cells (haESCs) can be established from uniparental blastocysts and possess only one set of chromosomes. Previous studies have shown that sperm or oocyte genome can be replaced by haESCs with or without manipulation of genomic imprinting for generation of mice. Recently, these remarkable semi-cloning methods have been applied for screening of key factors of mouse embryonic development. While haESCs have been applied as substitutes of gametic genomes, the fundamental mechanism how haESCs contribute to the genome of totipotent embryos is unclear. Here, we show the generation of fertile semi-cloned mice by injection of parthenogenetic haESCs (phaESCs) into oocytes after deletion of two differentially methylated regions (DMRs), the IG-DMR and H19-DMR. For characterizing the genome of semi-cloned embryos further, we establish ESC lines from semi-cloned blastocysts. We report that polyploid karyotypes are observed in semi-cloned ESCs (scESCs). Our results confirm that mitotically arrested phaESCs yield semi-cloned embryos and mice when the IG-DMR and H19-DMR are deleted. In addition, we highlight the occurrence of polyploidy that needs to be considered for further improving the development of semi-cloned embryos derived by haESC injection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Application of Mouse Parthenogenetic Haploid Embryonic Stem Cells as a Substitute of Sperm.J Vis Exp. 2020 Nov 19;(165). doi: 10.3791/61999. J Vis Exp. 2020. PMID: 33283788
-
Generation and application of mammalian haploid embryonic stem cells.J Intern Med. 2016 Sep;280(3):236-45. doi: 10.1111/joim.12503. Epub 2016 May 3. J Intern Med. 2016. PMID: 27138065 Review.
-
'Artificial spermatid'-mediated genome editing†.Biol Reprod. 2019 Sep 1;101(3):538-548. doi: 10.1093/biolre/ioz087. Biol Reprod. 2019. PMID: 31077288 Review.
-
Efficient Generation of Gene-Modified Mice by Haploid Embryonic Stem Cell-Mediated Semi-cloned Technology.Methods Mol Biol. 2017;1498:121-133. doi: 10.1007/978-1-4939-6472-7_8. Methods Mol Biol. 2017. PMID: 27709572
-
Temporal regulation of prenatal embryonic development by paternal imprinted loci.Sci China Life Sci. 2020 Jan;63(1):1-17. doi: 10.1007/s11427-019-9817-6. Epub 2019 Sep 17. Sci China Life Sci. 2020. PMID: 31564034
Cited by
-
In vitro gametogenesis: Towards competent oocytes: Limitations and future improvements for generating oocytes from pluripotent stem cells in culture.Bioessays. 2025 Jan;47(1):e2400106. doi: 10.1002/bies.202400106. Epub 2024 Nov 5. Bioessays. 2025. PMID: 39498732 Free PMC article. Review.
-
Epigenetic regulation limits competence of pluripotent stem cell-derived oocytes.EMBO J. 2023 Dec 1;42(23):e113955. doi: 10.15252/embj.2023113955. Epub 2023 Oct 18. EMBO J. 2023. PMID: 37850882 Free PMC article.
References
-
- Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308(5959):548–50. - PubMed
-
- Kawahara M, Wu Q, Takahashi N, Morita S, Yamada K, Ito M, et al. High-frequency generation of viable mice from engineered bi-maternal embryos. Nat Biotechnol. 2007;25(9):1045–50. - PubMed
-
- Kono T, Obata Y, Wu Q, Niwa K, Ono Y, Yamamoto Y, et al. Birth of parthenogenetic mice that can develop to adulthood. Nature. 2004;428(6985):860–4. - PubMed
-
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375(6526):34–9. - PubMed
-
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35(1):97–102. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

