Developmental, Dietary, and Geographical Impacts on Gut Microbiota of Red Swamp Crayfish (Procambarus clarkii)
- PMID: 32911609
- PMCID: PMC7565139
- DOI: 10.3390/microorganisms8091376
Developmental, Dietary, and Geographical Impacts on Gut Microbiota of Red Swamp Crayfish (Procambarus clarkii)
Abstract
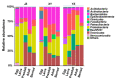
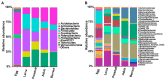
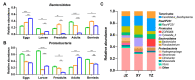
Red swamp crayfish (Procambarus clarkii) breeding is an important economic mainstay in Hubei province, China. However, information on the gut microbiota of the red swamp crayfish is limited. To address this issue, the effect of developmental stage, diet (fermented or non-fermented feed), and geographical location on the gut microbiota composition in the crayfish was studied via high-throughput 16S rRNA gene sequencing. The results revealed that the dominant phyla in the gut of the crayfish were Proteobacteria, Bacteroidetes,Firmicutes, Tenericutes, and RsaHF231. The alpha diversity showed a declining trend during development, and a highly comparable gut microbiota clustering was identified in a development-dependent manner. The results also revealed that development, followed by diet, is a better key driver for crayfish gut microbiota patterns than geographical location. Notably, the relative abundance of Bacteroidetes was significantly higher in the gut of the crayfish fed with fermented feed than those fed with non-fermented feed, suggesting the fermented feed can be important for the functions (e.g., polysaccharide degradation) of the gut microbiota. In summary, our results revealed the factors shaping gut microbiota of the crayfish and the importance of the fermented feed in crayfish breeding.
Keywords: Procambarus clarkii; fermented feed; gut microbiota; high-throughput sequencing; red swamp crayfish; shaping factors.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Gut microbiota of red swamp crayfish Procambarus clarkii in integrated crayfish-rice cultivation model.AMB Express. 2020 Jan 14;10(1):5. doi: 10.1186/s13568-019-0944-9. AMB Express. 2020. PMID: 31938890 Free PMC article.
-
The Role of Biogeography in Shaping Intestinal Flora and Influence on Fatty Acid Composition in Red Swamp Crayfish (Procambarus clarkii).Microb Ecol. 2023 Nov;86(4):3111-3127. doi: 10.1007/s00248-023-02298-4. Epub 2023 Oct 25. Microb Ecol. 2023. PMID: 37878052
-
Effects of Pelleted and Extruded Feed on Growth Performance, Intestinal Histology and Microbiota of Juvenile Red Swamp Crayfish (Procambarus clarkii).Animals (Basel). 2022 Aug 31;12(17):2252. doi: 10.3390/ani12172252. Animals (Basel). 2022. PMID: 36077973 Free PMC article.
-
Metagenomics Analysis Reveals Compositional and Functional Differences in the Gut Microbiota of Red Swamp Crayfish, Procambarus clarkii, Grown on Two Different Culture Environments.Front Microbiol. 2021 Oct 18;12:735190. doi: 10.3389/fmicb.2021.735190. eCollection 2021. Front Microbiol. 2021. PMID: 34733252 Free PMC article.
-
Red Swamp Crayfish (Procambarus clarkii) as a Growing Food Source: Opportunities and Challenges in Comprehensive Research and Utilization.Foods. 2024 Nov 25;13(23):3780. doi: 10.3390/foods13233780. Foods. 2024. PMID: 39682852 Free PMC article. Review.
Cited by
-
The hepatopancreas microbiome of velvet crab, Necora puber.Environ Microbiol Rep. 2024 Oct;16(5):e70014. doi: 10.1111/1758-2229.70014. Environ Microbiol Rep. 2024. PMID: 39354672 Free PMC article.
-
Isolation, Molecular Characterization, and Antimicrobial Resistance of Selected Culturable Bacteria From Crayfish (Procambarus clarkii).Front Microbiol. 2022 Jun 7;13:911777. doi: 10.3389/fmicb.2022.911777. eCollection 2022. Front Microbiol. 2022. PMID: 35747368 Free PMC article.
-
Effects of Different Sources of Culture Substrate on the Growth and Immune Performance of the Red Swamp Crayfish (Procambarus clarkii).Int J Mol Sci. 2023 Sep 14;24(18):14098. doi: 10.3390/ijms241814098. Int J Mol Sci. 2023. PMID: 37762400 Free PMC article.
-
Bacillus coagulans in Combination with Chitooligosaccharides Regulates Gut Microbiota and Ameliorates the DSS-Induced Colitis in Mice.Microbiol Spectr. 2022 Aug 31;10(4):e0064122. doi: 10.1128/spectrum.00641-22. Epub 2022 Jul 28. Microbiol Spectr. 2022. PMID: 35900082 Free PMC article.
-
The Microbiome Structure of a Rice-Crayfish Integrated Breeding Model and Its Association with Crayfish Growth and Water Quality.Microbiol Spectr. 2022 Apr 27;10(2):e0220421. doi: 10.1128/spectrum.02204-21. Epub 2022 Apr 6. Microbiol Spectr. 2022. PMID: 35384719 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

