ILRUN, a Human Plasma Lipid GWAS Locus, Regulates Lipoprotein Metabolism in Mice
- PMID: 32912065
- PMCID: PMC7644615
- DOI: 10.1161/CIRCRESAHA.120.317175
ILRUN, a Human Plasma Lipid GWAS Locus, Regulates Lipoprotein Metabolism in Mice
Abstract
Rationale: Single-nucleotide polymorphisms near the ILRUN (inflammation and lipid regulator with ubiquitin-associated-like and NBR1 [next to BRCA1 gene 1 protein]-like domains) gene are genome-wide significantly associated with plasma lipid traits and coronary artery disease (CAD), but the biological basis of this association is unknown.
Objective: To investigate the role of ILRUN in plasma lipid and lipoprotein metabolism.
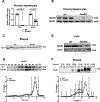
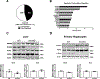
Methods and results: ILRUN encodes a protein that contains a ubiquitin-associated-like domain, suggesting that it may interact with ubiquitinylated proteins. We generated mice globally deficient for Ilrun and found they had significantly lower plasma cholesterol levels resulting from reduced liver lipoprotein production. Liver transcriptome analysis uncovered altered transcription of genes downstream of lipid-related transcription factors, particularly PPARα (peroxisome proliferator-activated receptor alpha), and livers from Ilrun-deficient mice had increased PPARα protein. Human ILRUN was shown to bind to ubiquitinylated proteins including PPARα, and the ubiquitin-associated-like domain of ILRUN was found to be required for its interaction with PPARα.
Conclusions: These findings establish ILRUN as a novel regulator of lipid metabolism that promotes hepatic lipoprotein production. Our results also provide functional evidence that ILRUN may be the casual gene underlying the observed genetic associations with plasma lipids at 6p21 in human.
Keywords: cholesterol; hepatocytes; lipids; lipoproteins; metabolism; ubiquitin.
Figures

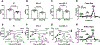



Comment in
-
Linking Lipid-Related Transcription and Cardiovascular Disease Through GWAS.Circ Res. 2020 Nov 6;127(11):1362-1364. doi: 10.1161/CIRCRESAHA.120.318221. Epub 2020 Nov 5. Circ Res. 2020. PMID: 33151796 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

