O- Fucosylation of ADAMTSL2 is required for secretion and is impacted by geleophysic dysplasia-causing mutations
- PMID: 32913123
- PMCID: PMC7667967
- DOI: 10.1074/jbc.RA120.014557
O- Fucosylation of ADAMTSL2 is required for secretion and is impacted by geleophysic dysplasia-causing mutations
Abstract
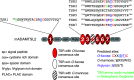
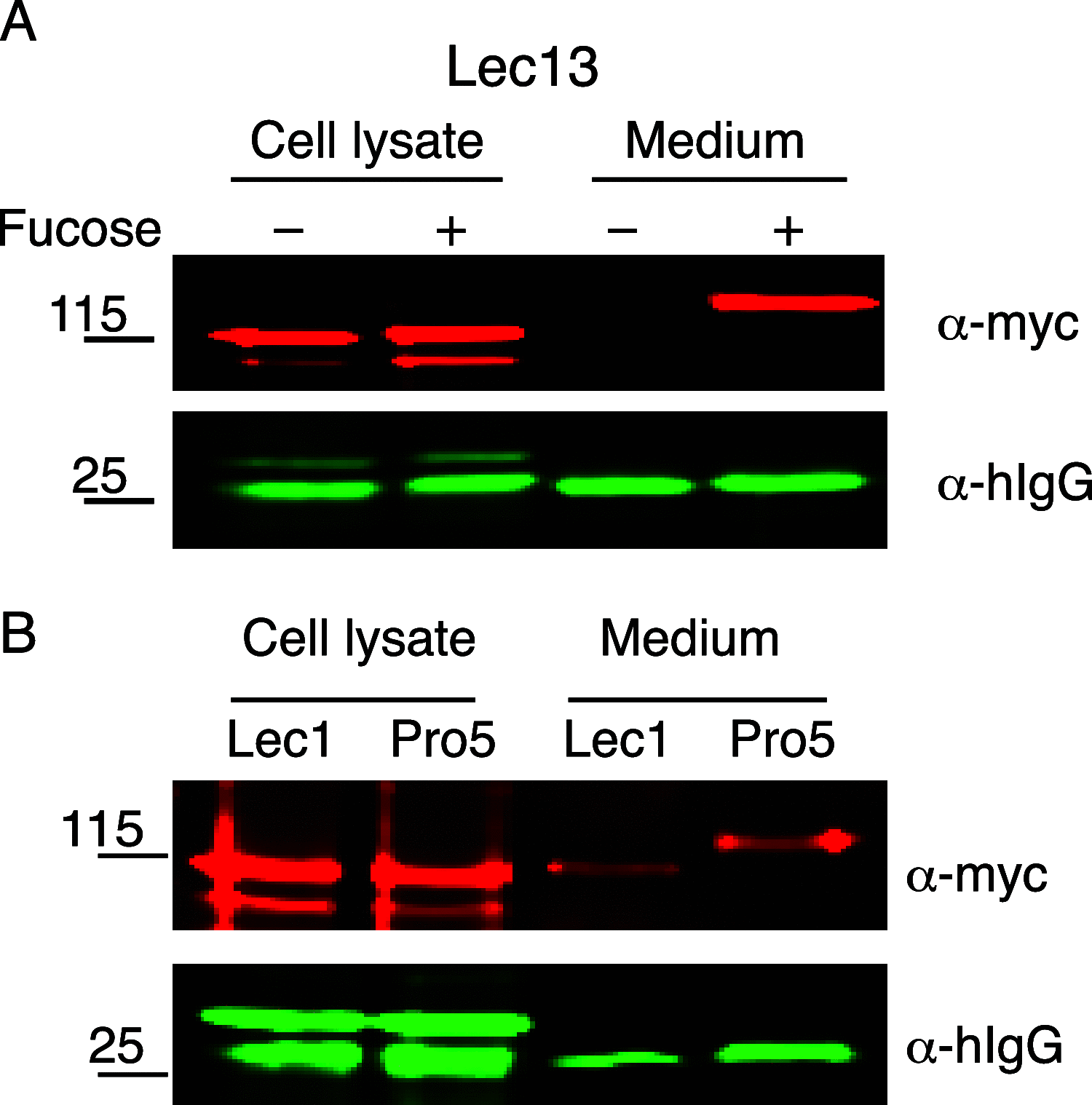
ADAMTSL2 mutations cause an autosomal recessive connective tissue disorder, geleophysic dysplasia 1 (GPHYSD1), which is characterized by short stature, small hands and feet, and cardiac defects. ADAMTSL2 is a matricellular protein previously shown to interact with latent transforming growth factor-β binding protein 1 and influence assembly of fibrillin 1 microfibrils. ADAMTSL2 contains seven thrombospondin type-1 repeats (TSRs), six of which contain the consensus sequence for O-fucosylation by protein O-fucosyltransferase 2 (POFUT2). O-fucose-modified TSRs are subsequently elongated to a glucose β1-3-fucose (GlcFuc) disaccharide by β1,3-glucosyltransferase (B3GLCT). B3GLCT mutations cause Peters Plus Syndrome (PTRPLS), which is characterized by skeletal defects similar to GPHYSD1. Several ADAMTSL2 TSRs also have consensus sequences for C-mannosylation. Six reported GPHYSD1 mutations occur within the TSRs and two lie near O-fucosylation sites. To investigate the effects of TSR glycosylation on ADAMTSL2 function, we used MS to identify glycan modifications at predicted consensus sequences on mouse ADAMTSL2. We found that most TSRs were modified with the GlcFuc disaccharide at high stoichiometry at O-fucosylation sites and variable mannose stoichiometry at C-mannosylation sites. Loss of ADAMTSL2 secretion in POFUT2-/- but not in B3GLCT-/- cells suggested that impaired ADAMTSL2 secretion is not responsible for skeletal defects in PTRPLS patients. In contrast, secretion was significantly reduced for ADAMTSL2 carrying GPHYSD1 mutations (S641L in TSR3 and G817R in TSR6), and S641L eliminated O-fucosylation of TSR3. These results provide evidence that abnormalities in GPHYSD1 patients with this mutation are caused by loss of O-fucosylation on TSR3 and impaired ADAMTSL2 secretion.
Keywords: ADAMTSL2; C-mannosylation; O-fucosylation; TSR; cell biology; extracellular matrix; geleophysic dysplasia; glycoprotein secretion; glycosylation; mass spectrometry.
© 2020 Zhang et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Similar articles
-
Analyzing the Effects of O-Fucosylation on Secretion of ADAMTS Proteins Using Cell-Based Assays.Methods Mol Biol. 2020;2043:25-43. doi: 10.1007/978-1-4939-9698-8_3. Methods Mol Biol. 2020. PMID: 31463900
-
ADAMTS9 and ADAMTS20 are differentially affected by loss of B3GLCT in mouse model of Peters plus syndrome.Hum Mol Genet. 2019 Dec 15;28(24):4053-4066. doi: 10.1093/hmg/ddz225. Hum Mol Genet. 2019. PMID: 31600785 Free PMC article.
-
Peters plus syndrome mutations affect the function and stability of human β1,3-glucosyltransferase.J Biol Chem. 2021 Jul;297(1):100843. doi: 10.1016/j.jbc.2021.100843. Epub 2021 May 28. J Biol Chem. 2021. PMID: 34058199 Free PMC article.
-
A report of three families with FBN1-related acromelic dysplasias and review of literature for genotype-phenotype correlation in geleophysic dysplasia.Eur J Med Genet. 2018 Apr;61(4):219-224. doi: 10.1016/j.ejmg.2017.11.018. Epub 2017 Nov 27. Eur J Med Genet. 2018. PMID: 29191498 Review.
-
Peters'-plus syndrome is a congenital disorder of glycosylation caused by a defect in the beta1,3-glucosyltransferase that modifies thrombospondin type 1 repeats.Ann Med. 2009;41(1):2-10. doi: 10.1080/07853890802301975. Ann Med. 2009. PMID: 18720094 Review.
Cited by
-
Protein O-Fucosyltransferases: Biological Functions and Molecular Mechanisms in Mammals.Molecules. 2025 Mar 26;30(7):1470. doi: 10.3390/molecules30071470. Molecules. 2025. PMID: 40286076 Free PMC article. Review.
-
O-fucosylation of thrombospondin type I repeats is dispensable for trafficking thrombospondin 1 to platelet secretory granules.Glycobiology. 2023 May 17;33(4):301-310. doi: 10.1093/glycob/cwad006. Glycobiology. 2023. PMID: 36721988 Free PMC article.
-
Dysregulation of cell migration by matrix metalloproteinases in geleophysic dysplasia.Sci Rep. 2025 Jun 6;15(1):19970. doi: 10.1038/s41598-025-04666-1. Sci Rep. 2025. PMID: 40481143 Free PMC article.
-
Case report: A homozygous ADAMTSL2 missense variant causes geleophysic dysplasia with high similarity to Weill-Marchesani syndrome.Front Genet. 2022 Sep 28;13:1014188. doi: 10.3389/fgene.2022.1014188. eCollection 2022. Front Genet. 2022. PMID: 36246610 Free PMC article.
-
A nonsense mutation in mouse Adamtsl2 causes uterine hypoplasia and an irregular estrous cycle.Mamm Genome. 2023 Dec;34(4):559-571. doi: 10.1007/s00335-023-10016-1. Epub 2023 Sep 1. Mamm Genome. 2023. PMID: 37656189 Free PMC article.
References
-
- Koo B. H., Le Goff C., Jungers K. A., Vasanji A., O'Flaherty J., Weyman C. M., and Apte S. S. (2007) ADAMTS-like 2 (ADAMTSL2) is a secreted glycoprotein that is widely expressed during mouse embryogenesis and is regulated during skeletal myogenesis. Matrix Biol. 26, 431–441 10.1016/j.matbio.2007.03.003 - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

