Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability
- PMID: 32913330
- PMCID: PMC7484789
- DOI: 10.1038/s41467-020-18306-x
Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability
Abstract
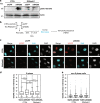
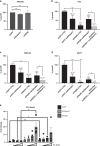
Collisions between the DNA replication machinery and co-transcriptional R-loops can impede DNA synthesis and are a major source of genomic instability in cancer cells. How cancer cells deal with R-loops to proliferate is poorly understood. Here we show that the ATP-dependent chromatin remodelling INO80 complex promotes resolution of R-loops to prevent replication-associated DNA damage in cancer cells. Depletion of INO80 in prostate cancer PC3 cells leads to increased R-loops. Overexpression of the RNA:DNA endonuclease RNAse H1 rescues the DNA synthesis defects and suppresses DNA damage caused by INO80 depletion. R-loops co-localize with and promote recruitment of INO80 to chromatin. Artificial tethering of INO80 to a LacO locus enabled turnover of R-loops in cis. Finally, counteracting R-loops by INO80 promotes proliferation and averts DNA damage-induced death in cancer cells. Our work suggests that INO80-dependent resolution of R-loops promotes DNA replication in the presence of transcription, thus enabling unlimited proliferation in cancers.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

