Clinical Value of RNA Sequencing-Based Classifiers for Prediction of the Five Conventional Breast Cancer Biomarkers: A Report From the Population-Based Multicenter Sweden Cancerome Analysis Network-Breast Initiative
- PMID: 32913985
- PMCID: PMC7446376
- DOI: 10.1200/PO.17.00135
Clinical Value of RNA Sequencing-Based Classifiers for Prediction of the Five Conventional Breast Cancer Biomarkers: A Report From the Population-Based Multicenter Sweden Cancerome Analysis Network-Breast Initiative
Abstract
Purpose: In early breast cancer (BC), five conventional biomarkers-estrogen receptor (ER), progesterone receptor (PgR), human epidermal growth factor receptor 2 (HER2), Ki67, and Nottingham histologic grade (NHG)-are used to determine prognosis and treatment. We aimed to develop classifiers for these biomarkers that were based on tumor mRNA sequencing (RNA-seq), compare classification performance, and test whether such predictors could add value for risk stratification.
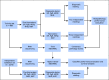
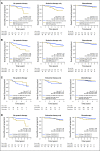
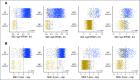
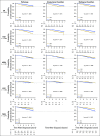
Methods: In total, 3,678 patients with BC were studied. For 405 tumors, a comprehensive multi-rater histopathologic evaluation was performed. Using RNA-seq data, single-gene classifiers and multigene classifiers (MGCs) were trained on consensus histopathology labels. Trained classifiers were tested on a prospective population-based series of 3,273 BCs that included a median follow-up of 52 months (Sweden Cancerome Analysis Network-Breast [SCAN-B], ClinicalTrials.gov identifier: NCT02306096), and results were evaluated by agreement statistics and Kaplan-Meier and Cox survival analyses.
Results: Pathologist concordance was high for ER, PgR, and HER2 (average κ, 0.920, 0.891, and 0.899, respectively) but moderate for Ki67 and NHG (average κ, 0.734 and 0.581). Concordance between RNA-seq classifiers and histopathology for the independent cohort of 3,273 was similar to interpathologist concordance. Patients with discordant classifications, predicted as hormone responsive by histopathology but non-hormone responsive by MGC, had significantly inferior overall survival compared with patients who had concordant results. This extended to patients who received no adjuvant therapy (hazard ratio [HR], 3.19; 95% CI, 1.19 to 8.57), or endocrine therapy alone (HR, 2.64; 95% CI, 1.55 to 4.51). For cases identified as hormone responsive by histopathology and who received endocrine therapy alone, the MGC hormone-responsive classifier remained significant after multivariable adjustment (HR, 2.45; 95% CI, 1.39 to 4.34).
Conclusion: Classification error rates for RNA-seq-based classifiers for the five key BC biomarkers generally were equivalent to conventional histopathology. However, RNA-seq classifiers provided added clinical value in particular for tumors determined by histopathology to be hormone responsive but by RNA-seq to be hormone insensitive.
© 2018 by American Society of Clinical Oncology.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center. Christian BruefferEmployment: SAGA Diagnostics ABJohan Vallon-ChristerssonNo relationship to discloseDorthe GrabauNo relationship to discloseAnna EhingerNo relationship to discloseJari HäkkinenNo relationship to discloseCecilia HegardtNo relationship to discloseJanne MalinaEmployment: Unilabs Honoraria: AstraZenecaYilun ChenNo relationship to disclosePär-Ola BendahlNo relationship to discloseJonas ManjerNo relationship to discloseMartin MalmbergNo relationship to discloseChrister LarssonHonoraria: Lilly (I) Research Funding: Diamyd Medical AB (I) Travel, Accommodations, Expenses: Lilly (I)Niklas LomanHonoraria: AstraZeneca Consulting or Advisory Role: AmgenLisa RydénResearch Funding: RocheÅke BorgHonoraria: Roche, AstraZeneca Travel, Accommodations, Expenses: Roche, AstraZenecaLao H. SaalEmployment: SAGA Diagnostics AB Leadership: SAGA Diagnostics AB Stock and Other Ownership Interests: SAGA Diagnostics AB Patents, Royalties, Other Intellectual Property: Patent filed for methods related to ultrasensitive quantification of nucleotide sequence variants.
Figures




References
-
- Gradishar WJ, Anderson BO, Blair SL, et al. : Breast cancer, version 3.2014. J Natl Compr Canc Netw 12:542-590, 2014 - PubMed
-
- Senkus E, Kyriakides S, Ohno S, et al. : Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 26:v8-v30, 2015 - PubMed
-
- Press MF, Sauter G, Bernstein L, et al. : Diagnostic evaluation of HER-2 as a molecular target: An assessment of accuracy and reproducibility of laboratory testing in large, prospective, randomized clinical trials. Clin Cancer Res 11:6598-6607, 2005 - PubMed
-
- Perez EA, Suman VJ, Davidson NE, et al. : HER2 testing by local, central, and reference laboratories in specimens from the North Central Cancer Treatment Group N9831 intergroup adjuvant trial. J Clin Oncol 24:3032-3038, 2006 - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

