Clinical Application of Circulating Tumor Cells and Circulating Tumor DNA in Uveal Melanoma
- PMID: 32913999
- PMCID: PMC7446501
- DOI: 10.1200/PO.17.00279
Clinical Application of Circulating Tumor Cells and Circulating Tumor DNA in Uveal Melanoma
Abstract
Purpose: To evaluate the feasibility of using circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA) for the management of uveal melanoma (UM).
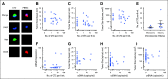
Patients and methods: Low-coverage whole-genome sequencing was used to determine somatic chromosomal copy number alterations (SCNAs) in primary UM tumors, ctDNA, and whole-genome amplified CTCs. CTCs were immunocaptured using an antimelanoma-associated chondroitin sulfate antibody conjugated to magnetic beads and immunostained for melanoma antigen recognised by T cells 1 (MART1)/glycoprotein 100 (gp100)/S100 calcium-binding protein β (S100β). ctDNA was quantified using droplet digital polymerase chain reaction assay for mutations in the GNAQ, GNA11, PLCβ4, and CYSLTR2 genes.
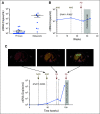
Results: SCNA analysis of CTCs and ctDNA isolated from a patient with metastatic UM showed good concordance with the enucleated primary tumor. In a cohort of 30 patients with primary UM, CTCs were detected in 58% of patients (one to 37 CTCs per 8 mL of blood), whereas only 26% of patients had detectable ctDNA (1.6 to 29 copies/mL). The presence of CTCs or ctDNA was not associated with tumor size or other prognostic markers. However, the frequent detection of CTCs in patients with early-stage UM supports a model in which CTCs can be used to derive tumor-specific SCNA relevant for prognosis. Monitoring of ctDNA after treatment of the primary tumor allowed detection of metastatic disease earlier than 18F-labeled fluorodeoxyglucose positron emission tomography in two patients.
Conclusion: The presence of CTCs in localized UM can be used to ascertain prognostic SCNA, whereas ctDNA can be used to monitor patients for early signs of metastatic disease. This study paves the way for the analysis of CTCs and ctDNA as a liquid biopsy that will assist with treatment decisions in patients with UM.
© 2018 by American Society of Clinical Oncology.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center. Aaron BeasleyNo relationship to discloseTimothy IsaacsTravel, Accommodations, Expenses: PfizerMuhammad A. KhattakHonoraria: MSD Oncology, Novartis, Merck Serono Consulting or Advisory Role: Bristol-Myers Squibb, Merck Serono Speakers' Bureau: Merck Serono, MSD Oncology, Novartis Research Funding: MSD Oncology Travel, Accommodations, Expenses: MSD Oncology, Amgen, Merck SeronoJames B. FreemanNo relationship to discloseRichard AllcockNo relationship to discloseFred K. ChenSpeakers' Bureau: Bayer HealthCare Pharmaceuticals Research Funding: Novartis (Inst) Travel, Accommodations, Expenses: Bayer HealthCare Pharmaceuticals, AllerganMichelle R. PereiraNo relationship to discloseKyle YauNo relationship to discloseJacqueline BentelNo relationship to discloseTersia VermeulenNo relationship to discloseLeslie CalapreNo relationship to discloseMichael MillwardConsulting or Advisory Role: Roche, Bristol-Myers Squibb, AstraZeneca, Merck Sharp & Dohme, Novartis, Boehringer Ingelheim Travel, Accommodations, Expenses: Roche, Merck Sharp & Dohme, Bristol-Myers Squibb, AstraZenecaMelanie R. ZimanResearch Funding: Merck Sharp & DohmeElin S. GrayResearch Funding: Merck Sharp & Dohme Patents, Royalties, Other Intellectual Property: Provisional patent on a blood test to detect melanoma based on auto-antibody detection Travel, Accommodations, Expenses: Bio-Rad Laboratories
Figures

References
-
- McLaughlin CC, Wu XC, Jemal A, et al. : Incidence of noncutaneous melanomas in the US. Cancer 103:1000-1007, 2005 - PubMed
-
- Blum ES, Yang J, Komatsubara KM, et al. : Clinical management of uveal and conjunctival melanoma. Oncology (Williston Park) 30:29-32, 34-43, 48, 2016 - PubMed
-
- Damato B, Dopierala JA, Coupland SE: Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res 16:6083-6092, 2010 - PubMed
LinkOut - more resources
Full Text Sources

