Prospective Clinical Validation of the InVisionFirst-Lung Circulating Tumor DNA Assay for Molecular Profiling of Patients With Advanced Nonsquamous Non-Small-Cell Lung Cancer
- PMID: 32914040
- PMCID: PMC7450945
- DOI: 10.1200/PO.18.00299
Prospective Clinical Validation of the InVisionFirst-Lung Circulating Tumor DNA Assay for Molecular Profiling of Patients With Advanced Nonsquamous Non-Small-Cell Lung Cancer
Abstract
Purpose: Guidelines advocate molecular profiling in the evaluation of advanced non-small-cell lung cancer (NSCLC) and support the use of plasma circulating tumor DNA (ctDNA)-based profiling for patients with insufficient tissue. Thorough prospective clinical validation studies of next-generation sequencing (NGS)-based ctDNA assays are lacking. We report the multicentered prospective clinical validation of the InVision ctDNA assay in patients with advanced untreated NSCLC.
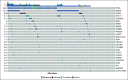
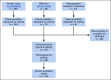
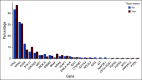
Methods: A total of 264 patients with untreated advanced NSCLC were prospectively recruited, and their plasma was analyzed using a ctDNA NGS assay for detection of genomic alterations in 36 commonly mutated genes. Tumor tissue was available in 178 patients for molecular profiling for comparison with plasma profiling. The remaining 86 patients were included to compare ctDNA profiles in patients with and without tissue for profiling.
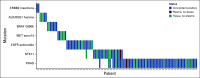
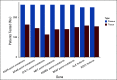
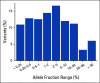
Results: Concordance of InVisionFirst with matched tissue profiling was 97.8%, with 82.9% positive predictive value, 98.5% negative predictive value, 70.6% sensitivity, and 99.2% specificity. Considering specific alterations in eight genes that most influence patient management, the positive predictive value was 97.8%, with 97.1% negative predictive value, 73.9% sensitivity, and 99.8% specificity. Across the entire study, 48 patients with actionable alterations were identified by ctDNA testing compared with only 38 by tissue testing. ctDNA NGS reported either an actionable alteration or an alteration generally considered mutually exclusive for such actionable changes in 53% of patients.
Conclusion: The liquid biopsy NGS assay demonstrated excellent concordance with tissue profiling in this multicenter, prospective, clinical validation study, with sensitivity and specificity equivalent to Food and Drug Administration-approved single-gene ctDNA assays. The use of plasma-based molecular profiling using NGS led to the detection of 26% more actionable alterations compared with standard-of-care tissue testing in this study.
© 2019 by American Society of Clinical Oncology.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center. Michael A. PritchettConsulting or Advisory Role: Biodesix, AstraZeneca, Medtronic, BodyVision Medical Speakers' Bureau: AstraZeneca, Medtronic, Boehringer Ingelheim, Actelion, United TherapeuticsD. Ross CamidgeHonoraria: Roche, G1 Therapeutics, Mersana, Takeda, AstraZeneca, Genoptix, Ignyta, Daiichi Sankyo, Hansoh, Lycera, Biothera, Revolution Medicines Research Funding: TakedaElke K. FriedmanLeadership: HCA Healthcare Stock and Other Ownership Interests: AbbVie (I) Honoraria: Gilead Sciences Consulting or Advisory Role: Gilead Sciences Travel, Accommodations, Expenses: Gilead SciencesSamir DaliaStock and Other Ownership Interests: 32nd Street Surgery Center (I) Consulting or Advisory Role: New Century Health Research Funding: Alnylam (Inst), Bristol-Myers Squibb (Inst), Calithera Biosciences (Inst), Helsinn Healthcare (Inst), Genentech (Inst), Celgene (Inst), Odonate Therapeutics (Inst)Katherine Baker-NeblettEmployment: Inivata Stock and Other Ownership Interests: GlaxoSmithKline Travel, Accommodations, Expenses: InivataVincent PlagnolEmployment: Inivata Stock and Other Ownership Interests: Inivata Patents, Royalties, Other Intellectual Property: Inivata patentsKaren D. HowarthEmployment: Inivata Stock and Other Ownership Interests: Inivata Research Funding: Inivata Patents, Royalties, Other Intellectual Property: Patent pendingGregory R. JonesEmployment: InivataNitzan RosenfeldEmployment: Storm Therapeutics (I) Leadership: Inivata Stock and Other Ownership Interests: Inivata, Mission Therapeutics (I) Research Funding: AstraZeneca (Inst) Patents, Royalties, Other Intellectual Property: Patents and patent applications relating to cancer classifications, detection, or analysis of microRNA and circulating tumor DNA, detection of rare sequence variants, applications in molecular diagnosticsClive D. MorrisEmployment: Inivata Leadership: Inivata Stock and Other Ownership Interests: InivataRamaswamy GovindanHonoraria: Genentech/AbbVie, AbbVie Consulting or Advisory Role: GlaxoSmithKline, Genentech, AbbVie, Celgene, AstraZeneca/MedImmune, Inivata, Merck Serono, Pfizer, Bristol-Myers Squibb, EMD Serono, Eli Lilly, Ignyta, Nektar, Phillips Gilmore Oncology, Jounce Therapeutics No other potential conflicts of interest were reported.
Figures

References
-
- Noone AM, Howlader N, Krapcho M et al. SEER cancer statistics review, 1975-2015. https://seer.cancer.gov/csr/1975_2015.
-
- Gutierrez ME, Choi K, Lanman RB, et al. Genomic profiling of advanced non-small cell lung cancer in community settings: Gaps and opportunities. Clin Lung Cancer. 2017;18:651–659. - PubMed
-
- Lokhandwala T, Bittoni MA, Dann RA, et al. Costs of diagnostic assessment for lung cancer: A Medicare claims analysis. Clin Lung Cancer. 2017;18:e27–e34. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

