Phylogenomic Resolution of Sea Spider Diversification through Integration of Multiple Data Classes
- PMID: 32915961
- PMCID: PMC7826184
- DOI: 10.1093/molbev/msaa228
Phylogenomic Resolution of Sea Spider Diversification through Integration of Multiple Data Classes
Abstract
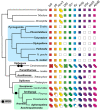
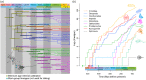
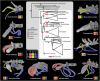
Despite significant advances in invertebrate phylogenomics over the past decade, the higher-level phylogeny of Pycnogonida (sea spiders) remains elusive. Due to the inaccessibility of some small-bodied lineages, few phylogenetic studies have sampled all sea spider families. Previous efforts based on a handful of genes have yielded unstable tree topologies. Here, we inferred the relationships of 89 sea spider species using targeted capture of the mitochondrial genome, 56 conserved exons, 101 ultraconserved elements, and 3 nuclear ribosomal genes. We inferred molecular divergence times by integrating morphological data for fossil species to calibrate 15 nodes in the arthropod tree of life. This integration of data classes resolved the basal topology of sea spiders with high support. The enigmatic family Austrodecidae was resolved as the sister group to the remaining Pycnogonida and the small-bodied family Rhynchothoracidae as the sister group of the robust-bodied family Pycnogonidae. Molecular divergence time estimation recovered a basal divergence of crown group sea spiders in the Ordovician. Comparison of diversification dynamics with other marine invertebrate taxa that originated in the Paleozoic suggests that sea spiders and some crustacean groups exhibit resilience to mass extinction episodes, relative to mollusk and echinoderm lineages.
Keywords: Pycnogonida; arthropods; diversification; mitogenome; ultraconserved.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Amato CG, Waugh DA, Feldmann RM, Schweitzer CE.. 2008. Effect of calcification on cuticle density in decapods: a key to lifestyle. J Crustac Biol. 28(4):587–595.
-
- Arabi J, Cruaud C, Couloux A, Hassanin A.. 2010. Studying sources of incongruence in arthropod molecular phylogenies: sea spiders (Pycnogonida) as a case study. C R Biol. 333(5):438–453. - PubMed
-
- Arango CP. 2002. Morphological phylogenetics of the sea spiders (Arthropoda: Pycnogonida). Org Divers Evol. 2(2):107–125.
-
- Arango CP, Wheeler WC.. 2007. Phylogeny of the sea spiders (Arthropoda, Pycnogonida) based on direct optimization of six loci and morphology. Cladistics 23(3):255–293. - PubMed
-
- Arnaud F, Bamber RN.. 1987. The biology of Pycnogonida. Adv Mar Biol. 24:1–95.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

