Circadian VIPergic Neurons of the Suprachiasmatic Nuclei Sculpt the Sleep-Wake Cycle
- PMID: 32916091
- PMCID: PMC7803671
- DOI: 10.1016/j.neuron.2020.08.001
Circadian VIPergic Neurons of the Suprachiasmatic Nuclei Sculpt the Sleep-Wake Cycle
Abstract
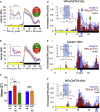
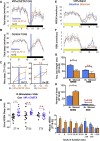
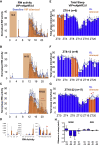
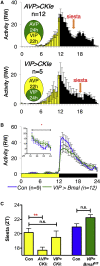
Although the mammalian rest-activity cycle is controlled by a "master clock" in the suprachiasmatic nucleus (SCN) of the hypothalamus, it is unclear how firing of individual SCN neurons gates individual features of daily activity. Here, we demonstrate that a specific transcriptomically identified population of mouse VIP+ SCN neurons is active at the "wrong" time of day-nighttime-when most SCN neurons are silent. Using chemogenetic and optogenetic strategies, we show that these neurons and their cellular clocks are necessary and sufficient to gate and time nighttime sleep but have no effect upon daytime sleep. We propose that mouse nighttime sleep, analogous to the human siesta, is a "hard-wired" property gated by specific neurons of the master clock to favor subsequent alertness prior to dawn (a circadian "wake maintenance zone"). Thus, the SCN is not simply a 24-h metronome: specific populations sculpt critical features of the sleep-wake cycle.
Keywords: alertness; circadian; napping; optogenetics; siesta; sleep; vasoactive intestinal polypeptide; wake maintenance.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

