Selective Lysosome Membrane Turnover Is Induced by Nutrient Starvation
- PMID: 32916093
- PMCID: PMC8337093
- DOI: 10.1016/j.devcel.2020.08.008
Selective Lysosome Membrane Turnover Is Induced by Nutrient Starvation
Abstract
Lysosome function is essential for cellular homeostasis, but quality-control mechanisms that maintain healthy lysosomes remain poorly characterized. Here, we developed a method to measure lysosome turnover and use this to identify a selective mechanism of membrane degradation that involves lipidation of the autophagy protein LC3 onto lysosomal membranes and the formation of intraluminal vesicles through microautophagy. This mechanism is induced in response to metabolic stress resulting from glucose starvation or by treatment with pharmacological agents that induce osmotic stress on lysosomes. Cells lacking ATG5, an essential component of the LC3 lipidation machinery, show reduced ability to regulate lysosome size and degradative capacity in response to activation of this mechanism. These findings identify a selective mechanism of lysosome membrane turnover that is induced by stress and uncover a function for LC3 lipidation in regulating lysosome size and activity through microautophagy.
Keywords: ATG5; LAP; LC3; ammonium; autophagy; glucose; glutamine; lysosome; metabolism; microautophagy.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



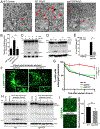
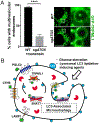
Comment in
-
Getting picky with the lysosome membrane.Autophagy. 2021 Apr;17(4):1034-1036. doi: 10.1080/15548627.2021.1877935. Epub 2021 Jan 26. Autophagy. 2021. PMID: 33499722 Free PMC article.
References
-
- Cai X, Xu Y, Cheung AK, Tomlinson RC, Alcazar-Roman A, Murphy L, Billich A, Zhang B, Feng Y, Klumpp M, et al. (2013). PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem Biol 20, 912–921. - PMC - PubMed
-
- Dikic I (2017). Proteasomal and Autophagic Degradation Systems. Annu Rev Biochem 86, 193–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

