Conserved Interferon-γ Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma
- PMID: 32916126
- PMCID: PMC7872287
- DOI: 10.1016/j.ccell.2020.08.005
Conserved Interferon-γ Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma
Erratum in
-
Conserved Interferon-γ Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma.Cancer Cell. 2021 Jan 11;39(1):122. doi: 10.1016/j.ccell.2020.11.015. Epub 2020 Dec 10. Cancer Cell. 2021. PMID: 33306984 Free PMC article. No abstract available.
Abstract
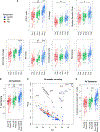
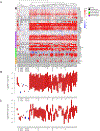
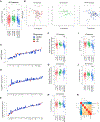
We analyze the transcriptome of baseline and on-therapy tumor biopsies from 101 patients with advanced melanoma treated with nivolumab (anti-PD-1) alone or combined with ipilimumab (anti-CTLA-4). We find that T cell infiltration and interferon-γ (IFN-γ) signaling signatures correspond most highly with clinical response to therapy, with a reciprocal decrease in cell-cycle and WNT signaling pathways in responding biopsies. We model the interaction in 58 human cell lines, where IFN-γ in vitro exposure leads to a conserved transcriptome response unless cells have IFN-γ receptor alterations. This conserved IFN-γ transcriptome response in melanoma cells serves to amplify the antitumor immune response. Therefore, the magnitude of the antitumor T cell response and the corresponding downstream IFN-γ signaling are the main drivers of clinical response or resistance to immune checkpoint blockade therapy.
Keywords: RNA-seq; anti-CTLA-4; anti-PD-1; biopsies; clinical trial; immune checkpoint blockade; immune exclusion; interferon-γ; resistance; response; transcriptomics.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests C.S.G. reports a pending patent on the use of PAK4 inhibitors. J.T. is currently an employee of Tango Therapeutics. M.O. reports no conflicts of interest. G.A.R. reports a pending patent on the use of PAK4 inhibitors. P.R.-M. is an employee and stockholder for Bristol-Myers Squibb. M.W.-R. is an employee and stockholder for Bristol-Myers Squibb. A.C. reports no conflicts of interest. E.M. reports no conflicts of interest. D.Y.T. reports no conflicts of interest. D.S.S. received honoraria from Genentech (speakers bureau) and NovMetaPharma (consultant). P.T. reports no conflicts of interest. Y.J.K. reports no conflicts of interest. C.P.S. has received payment for licensing a patent on non-viral T cell gene editing to Arsenal. K.M.C. is a shareholder in Geneoscopy LLC. A.V-C. reports no conflicts of interest. M.Q. reports no conflicts of interest. C.M. reports no conflicts of interest. J.J.L. discloses Data and Safety Monitoring Board: TTC Oncology; Scientific Advisory Board: 7 Hills, Actym, Alphamab Oncology, Kanaph, Mavu (now part of AbbVie), Onc.AI, Pyxis, Springbank, Tempest, Consultancy: Abbvie, Akrevia, Algios, Array, Astellas, Bayer, Bristol-Myers Squibb, Eisai, EMD Serono, Ideaya, Incyte, Janssen, Merck, Mersana, Novartis, PTx, RefleXion, Silicon, Tesaro, Vividion; Research Support: (all to institution for clinical trials unless noted) AbbVie, Agios (IIT), Array (IIT), Astellas, Bristol-Myers Squibb, CheckMate (SRA), Compugen, Corvus, EMD Serono, Evelo (SRA), Five Prime, FLX Bio, Genentech, Immatics, Immunocore, Incyte, Leap, MedImmune, Macrogenics, Necktar, Novartis, Palleon (SRA), Merck, Springbank, Tesaro, Tizona, Xencor; Travel: Akrevia, Bayer, Bristol-Myers Squibb, EMD Serono, Incyte, Janssen, Merck, Mersana, Novartis, Pyxis, RefleXion; Patents: (both provisional) serial no. 15/612,657 (Cancer Immunotherapy), PCT/US18/36052 (Microbiome Biomarkers for Anti-PD-1/PD-L1 Responsiveness: Diagnostic, Prognostic and Therapeutic Uses Thereof). J.D.W. is consultant for: Adaptive Biotech; Amgen; Apricity; Ascentage Pharma; Astellas; AstraZeneca; Bayer; Beigene; Bristol-Myers Squibb; Celgene; Eli Lilly; F Star; Imvaq; Kyowa Hakko Kirin; Linneaus; AstraZeneca; Merck; Neon Therapuetics; Polynoma; Psioxus; Recepta; Takara Bio; Trieza; Truvax; Serametrix; Surface Oncology; Syndax; Syntalogic. J.D.W. receives research support from: Bristol-Myers Squibb; AstraZeneca; Sephora. J.D.W. has Equity in: Tizona Pharmaceuticals; Adaptive Biotechnologies; Imvaq; Beigene; Linneaus. D.B.J. serves on advisory boards for Array BioPharma, BMS, Jansen, Merck, and Novartis, receives research funding from BMS and Incyte, and has a patent pending on using MHC-II as a biomarker for immunotherapy response. B.C. is a member of the speakers bureau for Regeneron and Sanofi. F.S.H. reports funding from Bristol-Myers Squibb to institution, during the conduct of the study; grants, personal fees and other from Bristol-Myers Squibb, personal fees from Merck, personal fees from EMD Serono, grants, personal fees and other from Novartis, personal fees from Takeda, personal fees from Surface, personal fees from Genentech/Roche, personal fees from Compass Therapeutics, personal fees from Apricity, personal fees from Bayer, personal fees from Aduro, personal fees from Partners Therapeutics, personal fees from Sanofi, personal fees from Pfizer, personal fees from Pionyr, personal fees from 7 Hills Pharma, personal fees from Verastem, personal fees from Torque, personal fees from Rheos, personal fees from Kairos, personal fees from Bicara, from Psioxus Therapeutics, personal fees from Amgen, other from Pieris Pharmaceutical, from Boston Pharmaceuticals, from Zumutor, outside the submitted work; In addition, Dr. Hodi has a patent for Methods for Treating MICA-Related Disorders (no. 20100111973) with royalties paid, a patent for Tumor Antigens and Uses Thereof (no. 7250291) issued, a patent for Angiopoiten-2 Biomarkers Predictive of Anti-immune checkpoint response (no. 20170248603) pending, a patent for Compositions and Methods for Identification, Assessment, Prevention, and Treatment of Melanoma using PD-L1 Isoforms (no. 20160340407) pending, a patent for Therapeutic Peptides (no. 20160046716) pending, a patent for Therapeutic Peptides (no. 20140004112) pending, a patent for Therapeutic Peptides (no. 20170022275) pending, a patent for Therapeutic Peptides (no. 20170008962) pending, a patent for Therapeutic Peptides patent no. 9402905 issued, a patent for Methods of Using Pembrolizumab and Trebananib pending, a patent for vaccine compositions and methods for restoring NKG2D pathway function against cancers patent no. 10279021 issued, a patent for antibodies that bind to MHC class I polypeptide-related sequence A patent no. 10106611 issued, and a patent for Anti-Galectin Antibody Biomarkers Predictive of Anti-Immune Checkpoint and Anti-Angiogenesis Responses publication no. 20170343552 pending. S.B. reports advisory board participation (with honorarium) from Genentech, EMD Serono, BMS and Sanofi-Genzyme; and research funding to his institution (University of Washington) from BMS, Oncosec, EMD Serono, Merck, NantKwest, Novartis, Exicure, Incyte, and Immune Design. W.S. discloses consulting fees from BMS, Merck, Novartis, Regeneron and research grant support from BMS, Merck, Novartis. W.J.U. serves on a compensated data and safety monitoring committee for Astra Zeneca, and reports institutional grants from BMS and Merck. C.L.S. discloses research funding to his University from Merck, Celldex, and GlaxoSmithKline; research support in kind to his University from Theraclion and 3M; Scientific Advisory Board role with Immatics (prior), and Curvac (planned); PI role for Polynoma with compensation to his University; and patent royalties as co-inventor of peptides for use in cancer vaccines (patents held by the UVA Licensing and Ventures Group). A.D. reports receiving honoraria from Nektar, Idera, Novartis, and Array BioPharma. J.B.A.G.H. Advisory roles: Aimm, Amgen, AZ, Bayer, BMS, Celsius, Gadeta, GSK, Immunocore, MSD, Merck Serono, Neon, Neogene, Novartis, Pfizer, Roche, Sanofi, Seattle Genetics, Vaximm. Grant support: Neon, BMS, MSD, Novartis. Stock options: Neogene Therapeutics. S.M.A. participated in advisory boards with Amgen, BMS, Merck Serono, MSD, Pierre-Fabre, Sanofi-Aventis and in educational events organized by BMS, MSD, Pierre-Fabre, Roche, Sanofi-Aventis. V.A. receives research funding from Bristol-Myers Squibb. D.M.P. and S.L.T. report stock and other ownership interests from Aduro Biotech, Compugen, DNAtrix, Dragonfly Therapeutics, Ervaxx, Five Prime Therapeutics, FLX Bio, Jounce Therapeutics, Potenza Therapeutics, Tizona Therapeutics, and WindMIL; consulting or advisory roles with AbbVie, Amgen, Bayer, Compugen, DNAtrix, Dragonfly Therapeutics, Dynavax, Ervaxx, Five Prime Therapeutics, FLX Bio, Immunocore, lmmunomic Therapeutics, Janssen Oncology, Medlmmune, Merck, Tizona Therapeutics, and Wind MIL; research funding from Bristol-Myers Squibb, Compugen, and Potenza Therapeutics; patents, royalties, and other intellectual property from Aduro Biotech, Bristol-Myers Squibb, and lmmunonomic Therapeutics; and travel, accommodations, and expenses from Bristol-Myers Squibb and Five Prime Therapeutics. V.E.V. is a founder of Delfi Diagnostics and Personal Genome Diagnostics, serves on the Board of Directors and as a consultant for both organizations, and owns Delfi Diagnostics and Personal Genome Diagnostics stock, which are subject to certain restrictions under university policy. In addition, Johns Hopkins University owns equity in Delfi Diagnostics and Personal Genome Diagnostics. V.E.V. is an advisor to Bristol-Myers Squibb, Genentech, Merck, and Takeda Pharmaceuticals. Within the last 5 years, V.E.V. has been an advisor to Daiichi Sankyo, Janssen Diagnostics, and Ignyta. These arrangements have been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies. D.E.S. reports no conflicts of interest. A.K. reports no conflicts of interest. A.R. has received honoraria from consulting with Amgen, Bristol-Myers Squibb, Chugai, Genentech, Merck, Novartis, Roche, and Sanofi, is or has been a member of the scientific advisory board and holds stock in Advaxis, Apricity, Arcus Biosciences, Bioncotech Therapeutics, Compugen, CytomX, Five Prime, FLX Bio, ImaginAb, Isoplexis, Kite-Gilead, Lutris Pharma, Merus, PACT Pharma, Rgenix, and Tango Therapeutics, has a reports a pending patent on the use of PAK4 inhibitors, has received research funding from Agilent and from Bristol-Myers Squibb through Stand Up to Cancer (SU2C), and has received payment for licensing a patent on non-viral T cell gene editing to Arsenal.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

