A Quantitative Proteome Map of the Human Body
- PMID: 32916130
- PMCID: PMC7575058
- DOI: 10.1016/j.cell.2020.08.036
A Quantitative Proteome Map of the Human Body
Abstract
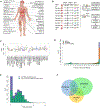
Determining protein levels in each tissue and how they compare with RNA levels is important for understanding human biology and disease as well as regulatory processes that control protein levels. We quantified the relative protein levels from over 12,000 genes across 32 normal human tissues. Tissue-specific or tissue-enriched proteins were identified and compared to transcriptome data. Many ubiquitous transcripts are found to encode tissue-specific proteins. Discordance of RNA and protein enrichment revealed potential sites of synthesis and action of secreted proteins. The tissue-specific distribution of proteins also provides an in-depth view of complex biological events that require the interplay of multiple tissues. Most importantly, our study demonstrated that protein tissue-enrichment information can explain phenotypes of genetic diseases, which cannot be obtained by transcript information alone. Overall, our results demonstrate how understanding protein levels can provide insights into regulation, secretome, metabolism, and human diseases.
Keywords: BBS syndrome; Leigh syndrome; TMT; branched-chain amino acid metabolism; mass spectrometry; protein and RNA correlation; quantitative proteomics; secretion; tissue-enriched or -specific proteins.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests M.P.S. is a cofounder and is on the scientific advisory board of Personalis, Filtircine, SensOmics, Qbio, January, Mirvie, Oralome, and Proteus. He is also on the scientific advisory board (SAB) of Genapsys and Jupiter. The other authors declare no competing interests.
Figures



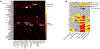
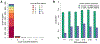
References
-
- Arany Z, and Neinast M (2018). Branched Chain Amino Acids in Metabolic Disease. Curr. Diab. Rep 18, 76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

