Advances in understanding the initiation of HIV-1 reverse transcription
- PMID: 32916568
- PMCID: PMC9973426
- DOI: 10.1016/j.sbi.2020.07.005
Advances in understanding the initiation of HIV-1 reverse transcription
Abstract
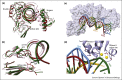
Many viruses, including Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Human Immunodeficiency Virus (HIV), use RNA as their genetic material. How viruses harness RNA structure and RNA-protein interactions to control their replication remains obscure. Recent advances in the characterization of HIV-1 reverse transcriptase, the enzyme that converts its single-stranded RNA genome into a double-stranded DNA copy, reveal how the reverse transcription complex evolves during initiation. Here we highlight these advances in HIV-1 structural biology and discuss how they are furthering our understanding of HIV and related ribonucleoprotein complexes implicated in viral disease.
Copyright © 2020. Published by Elsevier Ltd.
Figures


Similar articles
-
HIV-1 reverse transcription initiation: a potential target for novel antivirals?Virus Res. 2008 Jun;134(1-2):4-18. doi: 10.1016/j.virusres.2007.12.009. Epub 2008 Feb 6. Virus Res. 2008. PMID: 18255184 Review.
-
Dynamic Interplay of RNA and Protein in the Human Immunodeficiency Virus-1 Reverse Transcription Initiation Complex.J Mol Biol. 2018 Dec 7;430(24):5137-5150. doi: 10.1016/j.jmb.2018.08.029. Epub 2018 Sep 7. J Mol Biol. 2018. PMID: 30201267 Free PMC article.
-
Inhibition of both HIV-1 reverse transcription and gene expression by a cyclic peptide that binds the Tat-transactivating response element (TAR) RNA.PLoS Pathog. 2011 May;7(5):e1002038. doi: 10.1371/journal.ppat.1002038. Epub 2011 May 19. PLoS Pathog. 2011. PMID: 21625572 Free PMC article.
-
Stabilization of the U5-leader stem in the HIV-1 RNA genome affects initiation and elongation of reverse transcription.Nucleic Acids Res. 2000 Nov 1;28(21):4130-7. doi: 10.1093/nar/28.21.4130. Nucleic Acids Res. 2000. PMID: 11058109 Free PMC article.
-
Strand transfer events during HIV-1 reverse transcription.Virus Res. 2008 Jun;134(1-2):19-38. doi: 10.1016/j.virusres.2007.12.017. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279992 Review.
Cited by
-
Large-scale prediction shows that the dominant structure of the HIV-1 domain closed by the U5-AUG duplex contains the alternative SDa hairpin, and the domain variant without SD is rare.Virus Res. 2025 Jul;357:199581. doi: 10.1016/j.virusres.2025.199581. Epub 2025 May 15. Virus Res. 2025. PMID: 40381734 Free PMC article.
-
Let It Go: HIV-1 cis-Acting Repressive Sequences.J Virol. 2021 Jul 12;95(15):e0034221. doi: 10.1128/JVI.00342-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 33980600 Free PMC article. Review.
-
May I Help You with Your Coat? HIV-1 Capsid Uncoating and Reverse Transcription.Int J Mol Sci. 2024 Jun 28;25(13):7167. doi: 10.3390/ijms25137167. Int J Mol Sci. 2024. PMID: 39000271 Free PMC article. Review.
-
Chemical Control of CRISPR Gene Editing via Conditional Diacylation Crosslinking of Guide RNAs.Adv Sci (Weinh). 2023 Apr;10(10):e2206433. doi: 10.1002/advs.202206433. Epub 2023 Feb 3. Adv Sci (Weinh). 2023. PMID: 36737854 Free PMC article.
-
Bid Protein: A Participant in the Apoptotic Network with Roles in Viral Infections.Int J Mol Sci. 2025 Mar 7;26(6):2385. doi: 10.3390/ijms26062385. Int J Mol Sci. 2025. PMID: 40141030 Free PMC article. Review.
References
-
- WHO COVID19 statistics May 2020, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio..., on World Wide Web URL: http://www.WHO.int.
-
- WHO global health observatory data. www.who.int/gho/hiv/en/ on World Wide Web URL: http://www.WHO.int.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

