Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation
- PMID: 32916610
- PMCID: PMC7468592
- DOI: 10.1016/j.bios.2020.112572
Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation
Abstract
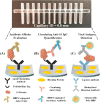
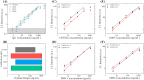
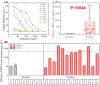
Convalescent serum with a high abundance of neutralization IgG is a promising therapeutic agent for rescuing COVID-19 patients in the critical stage. Knowing the concentration of SARS-CoV-2 S1-specific IgG is crucial in selecting appropriate convalescent serum donors. Here, we present a portable microfluidic ELISA technology for rapid (15 min), quantitative, and sensitive detection of anti-SARS-CoV-2 S1 IgG in human serum with only 8 μL sample volume. We first identified a humanized monoclonal IgG that has a high binding affinity and a relatively high specificity towards SARS-CoV-2 S1 protein, which can subsequently serve as the calibration standard of anti-SARS-CoV-2 S1 IgG in serological analyses. We then measured the abundance of anti-SARS-CoV-2 S1 IgG in 16 convalescent COVID-19 patients. Due to the availability of the calibration standard and the large dynamic range of our assay, we were able to identify "qualified donors" for convalescent serum therapy with only one fixed dilution factor (200 ×). Finally, we demonstrated that our technology can sensitively detect SARS-CoV-2 antigens (S1 and N proteins) with pg/mL level sensitivities in 40 min. Overall, our technology can greatly facilitate rapid, sensitive, and quantitative analysis of COVID-19 related markers for therapeutic, diagnostic, epidemiologic, and prognostic purposes.
Keywords: Antibody detection; COVID-19; Immunoassay; Microfluidics.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: The authors declare the following competing financial interest): M. K. K. O. and X. F. are co-founders of and have an equity interest in Optofluidic Bioassay, LLC.
Figures


References
-
- Arnold S., Ramjit R., Keng D., Kolchenko V., Teraoka I. Faraday Discuss. 2008;137:65–83. - PubMed
-
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., Bentlage A.E.H., van Haaren M.M., Guerra D., Burger J.A., Schermer E.E., Verheul K.D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M.J., Bijl T.P.L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N.A., Wiersinga W.J., Vidarsson G., Haagmans B.L., Ward A.B., de Bree G.J., Sanders R.W., van Gils M.J. Science. 2020;369(6504):643–650. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

