Maternal Origins and Haplotype Diversity of Seven Russian Goat Populations Based on the D-loop Sequence Variability
- PMID: 32916903
- PMCID: PMC7552281
- DOI: 10.3390/ani10091603
Maternal Origins and Haplotype Diversity of Seven Russian Goat Populations Based on the D-loop Sequence Variability
Abstract
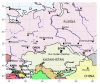
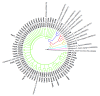
The territory of modern Russia lies on the crossroads of East and West and covers various geographical environments where diverse groups of local goats originated. In this work, we present the first study on the maternal origin of Russian local goats, including Altai Mountain (n = 9), Dagestan Downy (n = 18), Dagestan Local (n = 12), Dagestan Milk (n = 15), Karachaev (n = 21), Orenburg (n = 10), and Soviet Mohair (n = 7) breeds, based on 715 bp D-loop mitochondrial DNA (mtDNA) sequences. Saanen goats (n = 5) were used for comparison. Our findings reveal a high haplotype (HD = 0.843-1.000) and nucleotide diversity (π = 0.0112-0.0261). A total of 59 haplotypes were determined in the Russian goat breeds, in which all differed from the haplotypes of the Saanen goats. The haplotypes identified in Altai Mountain, Orenburg, Soviet Mohair, and Saanen goats were breed specific. Most haplotypes (56 of 59) were clustered together with samples belonging to haplogroup A, which was in accordance with the global genetic pattern of maternal origin seen in most goats worldwide. The haplotypes that were grouped together with rare haplogroups D and G were found in the Altai Mountain breed and haplogroup C was detected in the Soviet Mohair breed. Thus, our findings revealed that local goats might have been brought to Russia via various migration routes. In addition, haplotype sharing was found in aboriginal goat populations from overlapping regions, which might be useful information for their official recognition status.
Keywords: goat; haplogroup; livestock; local breeds; mtDNA; phylogeny.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
SNP-Based Genotyping Provides Insight Into the West Asian Origin of Russian Local Goats.Front Genet. 2021 Jul 1;12:708740. doi: 10.3389/fgene.2021.708740. eCollection 2021. Front Genet. 2021. PMID: 34276802 Free PMC article.
-
Characteristics of the Allele Pool and the Genetic Differentiation of Goats of Different Breeds and their Wild Relatives by Str-Markers.Arch Razi Inst. 2021 Nov 30;76(5):1351-1362. doi: 10.22092/ari.2021.355684.1709. eCollection 2021 Nov. Arch Razi Inst. 2021. PMID: 35355766 Free PMC article.
-
D-loop sequence mitochondrial DNA variability of Sarda goat and other goat breeds and populations reared in the Mediterranean area.J Anim Breed Genet. 2010 Oct;127(5):352-60. doi: 10.1111/j.1439-0388.2010.00863.x. J Anim Breed Genet. 2010. PMID: 20831559
-
Mitochondrial DNA D-loop sequence analysis reveals high variation and multiple maternal origins of indigenous Tanzanian goat populations.Ecol Evol. 2021 Nov 1;11(22):15961-15971. doi: 10.1002/ece3.8265. eCollection 2021 Nov. Ecol Evol. 2021. PMID: 34824803 Free PMC article.
-
Landmark native breed of the Orenburg goats: progress in its breeding and genetics and future prospects.Anim Biotechnol. 2023 Dec;34(9):5139-5154. doi: 10.1080/10495398.2022.2154221. Epub 2022 Dec 10. Anim Biotechnol. 2023. PMID: 36495096 Review.
Cited by
-
SNP-Based Genotyping Provides Insight Into the West Asian Origin of Russian Local Goats.Front Genet. 2021 Jul 1;12:708740. doi: 10.3389/fgene.2021.708740. eCollection 2021. Front Genet. 2021. PMID: 34276802 Free PMC article.
-
Ecogroups and maternal haplogroups reveal the ancestral origin of native Chinese goat populations based on the variation of mtDNA D-loop sequences.Ecol Evol. 2023 Aug 7;13(8):e10382. doi: 10.1002/ece3.10382. eCollection 2023 Aug. Ecol Evol. 2023. PMID: 37554396 Free PMC article.
-
Peeping into Mitochondrial Diversity of Andaman Goats: Unveils Possibility of Maritime Transport with Diversified Geographic Signaling.Genes (Basel). 2023 Mar 24;14(4):784. doi: 10.3390/genes14040784. Genes (Basel). 2023. PMID: 37107542 Free PMC article.
References
-
- Novopashina S.I., Sannikov M.Y., Khatataev S.A., Kuzmina T.N., Khmelevskaya G.N., Stepanova N.G., Tikhomirov A.I., Marinchenko T.E. Status and Perspective areas for Improving the Genetic Potential of Small Cattle: Scientific and Analytic Overview. Rosinformagrotekh; Moscow, Russia: 2019. pp. 27–39.
-
- Hajitov A.H., Stanishevskaya O.N., Safarov T.S. Biologicheskie i hozyajstvennye priznaki mestnyh koz. Izv. Spbgau. 2016;45:139–145. (In Russian)
-
- Grigoryan L.N., Hatataev S.A., Sverchkova S.V. Sostoyanie kozovodstva Rossijskoj Federacii i ego plemennoj bazy. In: Desyatov V.G., editor. Ezhegodnik po Plemennoj Rabote v Ovcevodstve i Kozovodstve v Hozyajstvah Rossijskoj Federacii (2005 god) Vserossijskij Nauchno-Issledovatel’skij Institut Plemennogo Dela; Lesnye Polyany, Russia: 2006. pp. 312–313. (In Russian)
-
- Dunin I.M., Amerhanov H.A., Safina G.F., Grigoryan L.N., Hatataev S.A., Hmelevskaya G.N., Pavlov M.B., Stepanova N.G. Kozovodstvo Rossii i ego plemennye resursy. In: Desyatov V.G., editor. Ezhegodnik po Plemennoj Rabote v Ovcevodstve i Kozovodstve v Hozyajstvah Rossijskoj Federacii (2019 god) Vserossijskij Nauchno-Issledovatel’skij Institut Plemennogo Dela; Lesnye Polyany, Russia: 2019. pp. 323–325. (In Russian)
-
- Musalaev K.K., Palaganova G.A., Abdullabekov R.A. Rezultaty nauchnykh issledovanii po ovtsevodstvu i kozovodstvu Dagestana. Gorn. Sel’skoe Hozyajstvo. 2015;2:121–124. (In Russian)
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

