Improved Detection of Antibodies against SARS-CoV-2 by Microsphere-Based Antibody Assay
- PMID: 32916926
- PMCID: PMC7555114
- DOI: 10.3390/ijms21186595
Improved Detection of Antibodies against SARS-CoV-2 by Microsphere-Based Antibody Assay
Abstract
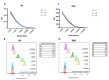
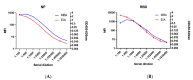
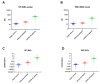
Currently available COVID-19 antibody tests using enzyme immunoassay (EIA) or immunochromatographic assay have variable sensitivity and specificity. Here, we developed and evaluated a novel microsphere-based antibody assay (MBA) for detecting immunoglobulin G (IgG) against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleoprotein (NP) and spike protein receptor binding domain (RBD). The seropositive cutoff value was set using a cohort of 294 anonymous serum specimens collected in 2018. The specificity was assessed using serum specimens collected from organ donors or influenza patients before 2020. Seropositive rate was determined among COVID-19 patients. Time-to-seropositivity and signal-to-cutoff (S/CO) ratio were compared between MBA and EIA. MBA had a specificity of 100% (93/93; 95% confidence interval (CI), 96-100%) for anti-NP IgG, 98.9% (92/93; 95% CI 94.2-100%) for anti-RBD IgG. The MBA seropositive rate for convalescent COVID-19 patients was 89.8% (35/39) for anti-NP IgG and 79.5% (31/39) for anti-RBD IgG. The time-to-seropositivity was shorter with MBA than EIA. MBA could better differentiate between COVID-19 patients and negative controls with higher S/CO ratio for COVID-19 patients, lower S/CO ratio with negative controls and fewer specimens in the equivocal range. MBA is robust, simple and is suitable for clinical microbiology laboratory for the accurate determination of anti-SARS-CoV-2 antibodies for diagnosis, serosurveillance, and vaccine trials.
Keywords: COVID-19; SARS-CoV-2; antibody assay; flow cytometry; serology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. - DOI - PMC - PubMed
-
- To K.K.-W., Cheng V.C.-C., Cai J., Chan K., Chen L., Wong L., Choi C.Y., Fong C.H., Ng A.C., Lu L., et al. Seroprevalence of SARS-CoV-2 in Hong Kong Special Administrative Region and our returnees evacuated from Hubei province of China: A multi-cohort study. Lancet Microbe. 2020;1:e111–e118. doi: 10.1016/S2666-5247(20)30053-7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

