Iron-Sulfur Cluster Complex Assembly in the Mitochondria of Arabidopsis thaliana
- PMID: 32917022
- PMCID: PMC7570111
- DOI: 10.3390/plants9091171
Iron-Sulfur Cluster Complex Assembly in the Mitochondria of Arabidopsis thaliana
Abstract
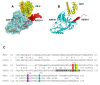
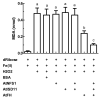
In plants, the cysteine desulfurase (AtNFS1) and frataxin (AtFH) are involved in the formation of Fe-S groups in mitochondria, specifically, in Fe and sulfur loading onto scaffold proteins, and the subsequent formation of the mature Fe-S cluster. We found that the small mitochondrial chaperone, AtISD11, and AtFH are positive regulators for AtNFS1 activity in Arabidopsis. Moreover, when the three proteins were incubated together, a stronger attenuation of the Fenton reaction was observed compared to that observed with AtFH alone. Using pull-down assays, we found that these three proteins physically interact, and sequence alignment and docking studies showed that several amino acid residues reported as critical for the interaction of their human homologous are conserved. Our results suggest that AtFH, AtNFS1 and AtISD11 form a multiprotein complex that could be involved in different stages of the iron-sulfur cluster (ISC) pathway in plant mitochondria.
Keywords: Arabidopsis; frataxin; mitochondria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Structural and functional studies of the mitochondrial cysteine desulfurase from Arabidopsis thaliana.Mol Plant. 2012 Sep;5(5):1001-10. doi: 10.1093/mp/sss037. Epub 2012 Apr 17. Mol Plant. 2012. PMID: 22511606
-
Altered levels of mitochondrial NFS1 affect cellular Fe and S contents in plants.Plant Cell Rep. 2019 Aug;38(8):981-990. doi: 10.1007/s00299-019-02419-9. Epub 2019 May 7. Plant Cell Rep. 2019. PMID: 31065779
-
The mitochondrial protein frataxin is essential for heme biosynthesis in plants.FEBS J. 2011 Feb;278(3):470-81. doi: 10.1111/j.1742-4658.2010.07968.x. Epub 2010 Dec 17. FEBS J. 2011. PMID: 21166997
-
Ancient and essential: the assembly of iron-sulfur clusters in plants.Trends Plant Sci. 2011 Apr;16(4):218-26. doi: 10.1016/j.tplants.2010.12.006. Epub 2011 Jan 21. Trends Plant Sci. 2011. PMID: 21257336 Review.
-
The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism.Biochim Biophys Acta. 2012 Sep;1823(9):1491-508. doi: 10.1016/j.bbamcr.2012.05.009. Epub 2012 May 15. Biochim Biophys Acta. 2012. PMID: 22609301 Review.
Cited by
-
Functional Relationships of Two NFU Proteins in Maintaining the Abundances of Mitochondrial Iron-Sulfur Proteins.Plant Direct. 2025 May 29;9(5):e70081. doi: 10.1002/pld3.70081. eCollection 2025 May. Plant Direct. 2025. PMID: 40443787 Free PMC article.
-
Iron in leaves: chemical forms, signalling, and in-cell distribution.J Exp Bot. 2022 Mar 15;73(6):1717-1734. doi: 10.1093/jxb/erac030. J Exp Bot. 2022. PMID: 35104334 Free PMC article. Review.
-
Plant-specific cochaperone SSR1 affects root elongation by modulating the mitochondrial iron-sulfur cluster assembly machinery.PLoS Genet. 2025 Feb 5;21(2):e1011597. doi: 10.1371/journal.pgen.1011597. eCollection 2025 Feb. PLoS Genet. 2025. PMID: 39908322 Free PMC article.
-
Structural and Functional Perspectives on Mitochondrial LYR-Domain Proteins in Plants.Physiol Plant. 2025 Jul-Aug;177(4):e70393. doi: 10.1111/ppl.70393. Physiol Plant. 2025. PMID: 40654039 Free PMC article.
-
Structural and Functional Characterization of CreFH1, the Frataxin Homolog from Chlamydomonas reinhardtii.Plants (Basel). 2022 Jul 26;11(15):1931. doi: 10.3390/plants11151931. Plants (Basel). 2022. PMID: 35893635 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

