A Single-Tube HNB-Based Loop-Mediated Isothermal Amplification for the Robust Detection of the Ostreid herpesvirus 1
- PMID: 32917059
- PMCID: PMC7555478
- DOI: 10.3390/ijms21186605
A Single-Tube HNB-Based Loop-Mediated Isothermal Amplification for the Robust Detection of the Ostreid herpesvirus 1
Abstract
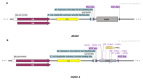
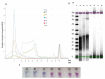
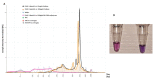
The Ostreid herpesvirus 1 species affects shellfish, contributing significantly to high economic losses during production. To counteract the threat related to mortality, there is a need for the development of novel point-of-care testing (POCT) that can be implemented in aquaculture production to prevent disease outbreaks. In this study, a simple, rapid and specific colorimetric loop-mediated isothermal amplification (LAMP) assay has been developed for the detection of Ostreid herpesvirus1 (OsHV-1) and its variants infecting Crassostrea gigas (C. gigas). The LAMP assay has been optimized to use hydroxynaphthol blue (HNB) for visual colorimetric distinction of positive and negative templates. The effect of an additional Tte UvrD helicase enzyme used in the reaction was also evaluated with an improved reaction time of 10 min. Additionally, this study provides a robust workflow for optimization of primers for uncultured viruses using designed target plasmid when DNA availability is limited.
Keywords: C. gigas; LAMP; OsHV-1; POCT; colorimetric; early warning detection; oyster.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Development of a loop-mediated isothermal amplification assay for rapid and sensitive detection of ostreid herpesvirus 1 DNA.J Virol Methods. 2010 Dec;170(1-2):30-6. doi: 10.1016/j.jviromet.2010.08.015. Epub 2010 Sep 8. J Virol Methods. 2010. PMID: 20813133
-
Detection of isothermally amplified ostreid herpesvirus 1 DNA in Pacific oyster (Crassostrea gigas) using a miniaturised electrochemical biosensor.Talanta. 2020 Jan 15;207:120308. doi: 10.1016/j.talanta.2019.120308. Epub 2019 Sep 2. Talanta. 2020. PMID: 31594570
-
Rapid visual detection of Aeromonas salmonicida by loop-mediated isothermal amplification with hydroxynaphthol blue dye.J Fish Dis. 2021 Dec;44(12):1993-2001. doi: 10.1111/jfd.13513. Epub 2021 Aug 19. J Fish Dis. 2021. PMID: 34411329
-
Rapid visual detection of Vibrio parahaemolyticus in seafood samples by loop-mediated isothermal amplification with hydroxynaphthol blue dye.World J Microbiol Biotechnol. 2020 May 10;36(5):76. doi: 10.1007/s11274-020-02851-0. World J Microbiol Biotechnol. 2020. PMID: 32390085
-
An optimized visual loop mediated isothermal amplification assay for efficient detection of minute virus of mice with hydroxynaphthol blue dye.J Virol Methods. 2022 Oct;308:114575. doi: 10.1016/j.jviromet.2022.114575. Epub 2022 Jul 5. J Virol Methods. 2022. PMID: 35798197
Cited by
-
Integrated microfluidic cartridge for rapid and colorimetric detection of Mycoplasma pneumoniae.Mikrochim Acta. 2025 Aug 6;192(9):559. doi: 10.1007/s00604-025-07377-6. Mikrochim Acta. 2025. PMID: 40767967
-
Improving Performance of a SARS-CoV-2 RT-LAMP Assay for Use With a Portable Isothermal Fluorimeter: Towards a Point-of-Care Molecular Testing Strategy.J Biomol Tech. 2021 Sep;32(3):180-185. doi: 10.7171/jbt.21-3203-013. J Biomol Tech. 2021. PMID: 35027875 Free PMC article.
-
Sensitivity Validation of EWOD Devices for Diagnosis of Early Mortality Syndrome (EMS) in Shrimp Using Colorimetric LAMP-XO Technique.Sensors (Basel). 2021 Apr 30;21(9):3126. doi: 10.3390/s21093126. Sensors (Basel). 2021. PMID: 33946302 Free PMC article.
-
A colorimetric hydroxy naphthol blue based loop-mediated isothermal amplification detection assay targeting the β-tubulin locus of Sarocladium oryzae infecting rice seed.Front Plant Sci. 2022 Nov 21;13:1077328. doi: 10.3389/fpls.2022.1077328. eCollection 2022. Front Plant Sci. 2022. PMID: 36479512 Free PMC article.
-
Sex identification of sun conure (Aratinga solstitialis) using loop-mediated isothermal amplification of W and Z spindlin chromosomes.Vet World. 2024 Sep;17(9):2000-2007. doi: 10.14202/vetworld.2024.2000-2007. Epub 2024 Sep 8. Vet World. 2024. PMID: 39507791 Free PMC article.
References
-
- Pellet P.E., Roizman B. The family Herpesviridae: A brief introduction. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 2479–2499.
-
- Hellberg T., Paßvogel L., Schulz K.S., Klupp B.G., Mettenleiter T.C. Nuclear egress of Herpesviruses: The prototypic vesicular nucleocytoplasmic transport. Adv. Virus Res. 2016;94:81–140. - PubMed
-
- Lionel D., Guyader T., Tourbiez D., Pépin J.F. Is horizontal transmission of the Ostreid herpesvirus OsHV-1 in Crassostrea gigas affected by unselected or selected survival status in adults to juveniles? Aquaculture. 2013;408–409:51–57. doi: 10.1016/j.aquaculture.2013.05.025. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

