Hepatocyte-like cells derived from human induced pluripotent stem cells using small molecules: implications of a transcriptomic study
- PMID: 32917265
- PMCID: PMC7488531
- DOI: 10.1186/s13287-020-01914-1
Hepatocyte-like cells derived from human induced pluripotent stem cells using small molecules: implications of a transcriptomic study
Abstract
Background: Hepatocyte-like cells (HLCs) derived from human induced pluripotent stem cells (iPSCs) hold great promise in toxicological applications as well as in regenerative medicine. Previous efforts on hepatocyte differentiation have mostly relied on the use of growth factors (GFs) to recapitulate developmental signals under in vitro conditions. Recently, the use of small molecules (SMs) has emerged as an attractive tool to induce cell fate transition due to its superiority in terms of both quality and cost. However, HLCs derived using SMs have not been well characterized, especially on the transcriptome level.
Methods: HLCs were differentiated from human iPSCs using a protocol that only involves SMs and characterized by transcriptomic analysis using whole genome microarrays.
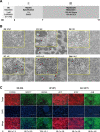
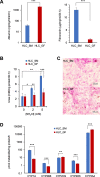
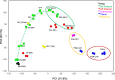
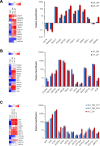
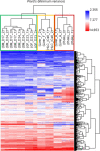
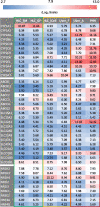
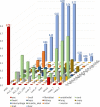
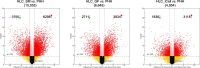
Results: HLCs derived using the SM protocol (HLC_SM) displayed specific hepatic marker expression and demonstrated key hepatic functions. Transcriptomic analysis of the SM-driven differentiation defined a hepatocyte differentiation track and characterized the expression of some key marker genes in major stages of hepatocyte differentiation. In addition, HLC_SM were scored with CellNet, a bioinformatics tool quantifying how closely engineered cell populations resemble their target cell type, and compared to primary human hepatocytes (PHHs), adult liver tissue, fetal liver tissue, HLCs differentiated using GFs (HLC_GF), and commercially available HLCs. Similar to HLC_GF, HLC_SM displayed a mixed phenotype of fetal and adult hepatocytes and had relatively low expression of metabolic enzymes, transporters, and nuclear receptors compared to PHHs. Finally, the differentially expressed genes in HLC_SM compared to HLC_GF and to PHHs were analyzed to identify pathways and upstream transcription regulators which could potentially be manipulated to improve the differentiation of HLCs.
Conclusions: Overall, the present study demonstrated the usefulness of the SM-based hepatocyte differentiation method, offered new insights into the molecular basis of hepatogenesis and associated gene regulation, and suggested ways for further improvements in hepatocyte differentiation in order to obtain more mature HLCs that could be used in toxicological studies.
Keywords: Hepatocyte differentiation; Hepatocyte-like cells; Induced pluripotent stem cells; Microarray; Small molecules; Transcriptomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Analysis of differentially expressed genes among human hair follicle-derived iPSCs, induced hepatocyte-like cells, and primary hepatocytes.Stem Cell Res Ther. 2018 Aug 9;9(1):211. doi: 10.1186/s13287-018-0940-z. Stem Cell Res Ther. 2018. PMID: 30092828 Free PMC article.
-
A comprehensive transcriptomic comparison of hepatocyte model systems improves selection of models for experimental use.Commun Biol. 2022 Oct 14;5(1):1094. doi: 10.1038/s42003-022-04046-9. Commun Biol. 2022. PMID: 36241695 Free PMC article.
-
Liver biopsy derived induced pluripotent stem cells provide unlimited supply for the generation of hepatocyte-like cells.PLoS One. 2019 Aug 29;14(8):e0221762. doi: 10.1371/journal.pone.0221762. eCollection 2019. PLoS One. 2019. PMID: 31465481 Free PMC article.
-
Phenotypical, functional and transcriptomic comparison of two modified methods of hepatocyte differentiation from human induced pluripotent stem cells.Biomed Rep. 2022 May;16(5):43. doi: 10.3892/br.2022.1526. Epub 2022 Mar 23. Biomed Rep. 2022. PMID: 35371477 Free PMC article.
-
Current status of hepatocyte-like cell therapy from stem cells.Surg Today. 2021 Mar;51(3):340-349. doi: 10.1007/s00595-020-02092-6. Epub 2020 Aug 4. Surg Today. 2021. PMID: 32754843 Review.
Cited by
-
Elimination of Reprogramming Transgenes Facilitates the Differentiation of Induced Pluripotent Stem Cells into Hepatocyte-like Cells and Hepatic Organoids.Biology (Basel). 2022 Mar 23;11(4):493. doi: 10.3390/biology11040493. Biology (Basel). 2022. PMID: 35453693 Free PMC article.
-
5-Aminolevulinate improves metabolic recovery and cell survival of the liver following cold preservation.Theranostics. 2022 Mar 21;12(6):2908-2927. doi: 10.7150/thno.69446. eCollection 2022. Theranostics. 2022. PMID: 35401816 Free PMC article.
-
Standard Protocols for Characterising Primary and In Vitro-Generated Human Hepatocytes.J Cell Mol Med. 2025 Feb;29(3):e70390. doi: 10.1111/jcmm.70390. J Cell Mol Med. 2025. PMID: 39910642 Free PMC article. Review.
-
Hepatocytes derived from human induced pluripotent stem cells: Towards establishing an in vitro model for assessing population variability in hepatotoxicity testing.Curr Res Toxicol. 2025 May 15;8:100238. doi: 10.1016/j.crtox.2025.100238. eCollection 2025. Curr Res Toxicol. 2025. PMID: 40496982 Free PMC article.
-
Comparative analysis of small molecule and growth factor-derived human induced pluripotent stem cell-derived hepatocyte-like cells.Front Cell Dev Biol. 2025 Jun 26;13:1594340. doi: 10.3389/fcell.2025.1594340. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40641603 Free PMC article.
References
-
- Hewitt NJ, Lechon MJ, Houston JB, Hallifax D, Brown HS, Maurel P, et al. Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab Rev. 2007;39(1):159–234. doi: 10.1080/03602530601093489. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

