Premeiotic, 24-Nucleotide Reproductive PhasiRNAs Are Abundant in Anthers of Wheat and Barley But Not Rice and Maize
- PMID: 32917771
- PMCID: PMC7608162
- DOI: 10.1104/pp.20.00816
Premeiotic, 24-Nucleotide Reproductive PhasiRNAs Are Abundant in Anthers of Wheat and Barley But Not Rice and Maize
Abstract
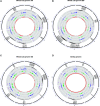
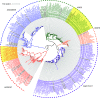
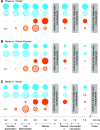
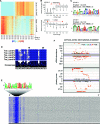
Two classes of premeiotic (21-nucleotides [nt]) and meiotic (24-nt) phased small interfering RNAs (phasiRNAs) and their patterns of accumulation have been described in maize (Zea mays) and rice (Oryza sativa) anthers. Their precise function remains unclear, but studies have shown that they support male fertility. The important role of phasiRNAs in anthers underpins our present study to characterize these small RNAs in wheat (Triticum aestivum) and barley (Hordeum vulgare) anthers. We staged anthers at every 0.2 mm of development for one wheat and two barley varieties. We isolated premeiotic (0.2, 0.4, and 0.6 mm), meiotic (0.8, 1.0, and 1.4 mm), and postmeiotic (1.8 mm) anthers, for which we then investigated accumulation patterns of RNAs, including reproductive phasiRNAs. We annotated a total of 12,821 and 2,897 PHAS loci in the wheat and barley genomes, respectively. By comparing the total number of PHAS loci in genomes of maize, rice, barley, and wheat, we identified an expansion of reproductive PHAS loci in the genomes of Poaceae subfamilies from Panicoideae to Oryzoideae and to Poideae. In addition to the two classes of premeiotic (21-nt) and meiotic (24-nt) phasiRNAs, previously described in maize and rice anthers, we characterized a group of 24-nt phasiRNAs that accumulate in premeiotic anthers. The absence of premeiotic 24-nt phasiRNAs in maize and rice suggests a divergence in grass species of the Poideae subfamily. Additionally, we performed a gene coexpression analysis describing the regulation of phasiRNA biogenesis in wheat and barley anthers. We highlight Argonaute 9 (AGO9) and Argonaute 6 (AGO6) as candidate binding partners of premeiotic and meiotic 24-nt phasiRNAs, respectively.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

