Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor
- PMID: 32917884
- PMCID: PMC7486399
- DOI: 10.1038/s41467-020-18319-6
Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor
Erratum in
-
Author Correction: Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor.Nat Commun. 2021 May 14;12(1):2996. doi: 10.1038/s41467-021-23498-x. Nat Commun. 2021. PMID: 33990607 Free PMC article. No abstract available.
Abstract
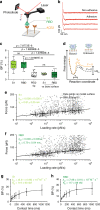
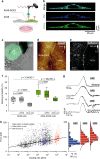
Study of the interactions established between the viral glycoproteins and their host receptors is of critical importance for a better understanding of virus entry into cells. The novel coronavirus SARS-CoV-2 entry into host cells is mediated by its spike glycoprotein (S-glycoprotein), and the angiotensin-converting enzyme 2 (ACE2) has been identified as a cellular receptor. Here, we use atomic force microscopy to investigate the mechanisms by which the S-glycoprotein binds to the ACE2 receptor. We demonstrate, both on model surfaces and on living cells, that the receptor binding domain (RBD) serves as the binding interface within the S-glycoprotein with the ACE2 receptor and extract the kinetic and thermodynamic properties of this binding pocket. Altogether, these results provide a picture of the established interaction on living cells. Finally, we test several binding inhibitor peptides targeting the virus early attachment stages, offering new perspectives in the treatment of the SARS-CoV-2 infection.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

