Protrudin and PDZD8 contribute to neuronal integrity by promoting lipid extraction required for endosome maturation
- PMID: 32917905
- PMCID: PMC7486383
- DOI: 10.1038/s41467-020-18413-9
Protrudin and PDZD8 contribute to neuronal integrity by promoting lipid extraction required for endosome maturation
Abstract
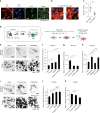
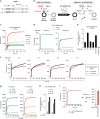
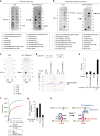
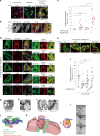
Endosome maturation depends on membrane contact sites (MCSs) formed between endoplasmic reticulum (ER) and endolysosomes (LyLEs). The mechanism underlying lipid supply for this process and its pathophysiological relevance remains unclear, however. Here, we identify PDZD8-the mammalian ortholog of a yeast ERMES subunit-as a protein that interacts with protrudin, which is located at ER-LyLE MCSs. Protrudin and PDZD8 promote the formation of ER-LyLE MCSs, and PDZD8 shows the ability to extract various lipids from the ER. Overexpression of both protrudin and PDZD8 in HeLa cells, as well as their depletion in mouse primary neurons, impairs endosomal homeostasis by inducing the formation of abnormal large vacuoles reminiscent of those apparent in spastin- or REEP1-deficient neurons. The protrudin-PDZD8 system is also essential for the establishment of neuronal polarity. Our results suggest that protrudin and PDZD8 cooperatively promote endosome maturation by mediating ER-LyLE tethering and lipid extraction at MCSs, thereby maintaining neuronal polarity and integrity.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Helle SC, et al. Organization and function of membrane contact sites. Biochim. Biophys. Acta. 2013;1833:2526–2541. - PubMed
-
- Salvador-Gallego R, Hoyer MJ, Voeltz GK. SnapShot: functions of endoplasmic reticulum membrane contact sites. Cell. 2017;171:1224–1224. - PubMed
-
- Saheki Y, De Camilli P. Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 2017;86:659–684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

