Genetic variation regulates opioid-induced respiratory depression in mice
- PMID: 32917924
- PMCID: PMC7486296
- DOI: 10.1038/s41598-020-71804-2
Genetic variation regulates opioid-induced respiratory depression in mice
Abstract
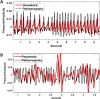
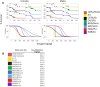
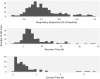
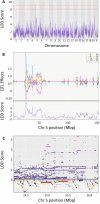
In the U.S., opioid prescription for treatment of pain nearly quadrupled from 1999 to 2014. The diversion and misuse of prescription opioids along with increased use of drugs like heroin and fentanyl, has led to an epidemic in addiction and overdose deaths. The most common cause of opioid overdose and death is opioid-induced respiratory depression (OIRD), a life-threatening depression in respiratory rate thought to be caused by stimulation of opioid receptors in the inspiratory-generating regions of the brain. Studies in mice have revealed that variation in opiate lethality is associated with strain differences, suggesting that sensitivity to OIRD is genetically determined. We first tested the hypothesis that genetic variation in inbred strains of mice influences the innate variability in opioid-induced responses in respiratory depression, recovery time and survival time. Using the founders of the advanced, high-diversity mouse population, the Diversity Outbred (DO), we found substantial sex and genetic effects on respiratory sensitivity and opiate lethality. We used DO mice treated with morphine to map quantitative trait loci for respiratory depression, recovery time and survival time. Trait mapping and integrative functional genomic analysis in GeneWeaver has allowed us to implicate Galnt11, an N-acetylgalactosaminyltransferase, as a gene that regulates OIRD.
Conflict of interest statement
B.F.O. and K.D.D. are co-founders and co-owners of Signal Solutions that manufactures and sells the Piezoelectric technology utilized in this study.
Figures




References
-
- Boscarino JA, et al. Risk factors for drug dependence among out-patients on opioid therapy in a large US health-care system. Addiction. 2010;105:1776–1782. - PubMed
-
- Cicero TJ, Kasper ZA, Ellis MS. Increased use of heroin as an initiating opioid of abuse: Further considerations and policy implications. Addict. Behav. 2018;87:267–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

