LncRNA HOXA-AS3 promotes the malignancy of glioblastoma through regulating miR-455-5p/USP3 axis
- PMID: 32918360
- PMCID: PMC7579690
- DOI: 10.1111/jcmm.15788
LncRNA HOXA-AS3 promotes the malignancy of glioblastoma through regulating miR-455-5p/USP3 axis
Abstract
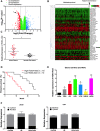
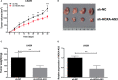
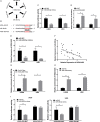
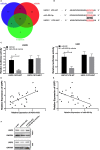
Our objective was to determine the molecular mechanisms by which lncRNA HOXA-AS3 regulates the biological behaviour of glioblastoma multiforme (GBM). We used an lncRNA microarray assay to identify GBM-related lncRNA expression profiles. Qrt-PCR was used to survey the levels of expression of long non-coding RNA (lncRNA) HOXA-AS3 and the target gene. Dual-luciferase reporter assays were used to investigate the interaction of lncRNA HOXA-AS3, the target gene and miRNA. Western blot analysis was used to examine the expression of USP3 and epithelial-mesenchymal transition (EMT) genes. The MTT assay, transwell assay and wound healing assay were used to analyse the effects of lncRNA HOXA-AS3 on GBM cell viability, mobility and invasiveness, respectively. Our results showed that lncRNA HOXA-AS3 was significantly up-regulated in GBM cells and could promote GBM cell proliferation, invasion and migration in vitro and in vivo. HOXA-AS was found to be associated with poor survival prognosis in glioma patients. The dual-luciferase reporter assay also revealed that lncRNA HOXA-AS3 acts as a mir-455-5p sponge by up-regulating USP3 expression to promote GBM progression. Western blot analysis showed that lncRNA HOXA-AS3 could up-regulate EMT-related gene expression in GBM. Experiments showed mir-455-5p could rescue the effect of lncRNA HOXA-AS3 on cell proliferation and invasion. The newly identified HOXA-AS3/mir-455-5p/USP3 pathway offers important clues to understanding the key mechanisms underlying the action of lncRNA HOXA-AS3 in glioblastoma.
Keywords: USP3; competing endogenous RNA; glioblastoma; lncRNA HOXA-AS3; miR-455-5p.
© 2020 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors confirm that there are no conflicts of interest.
Figures

References
-
- Tian Y, Zheng Y, Dong X. AGAP2‐AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J Cell Biochem. 2019;120(6):9056‐9062. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

