Differential and unique patterns of synaptic miRNA expression in dorsolateral prefrontal cortex of depressed subjects
- PMID: 32919404
- PMCID: PMC8115313
- DOI: 10.1038/s41386-020-00861-y
Differential and unique patterns of synaptic miRNA expression in dorsolateral prefrontal cortex of depressed subjects
Abstract
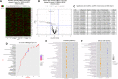
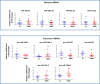
Altered synaptic plasticity is often associated with major depressive disorder (MDD). Disease-associated changes in synaptic functions are tightly correlated with altered microRNA (miRNA) expression. Here, we examined the role of miRNAs and their functioning at the synapse in MDD by examining miRNA processing machinery at synapse and sequencing miRNAs and analyzing their functions in synaptic and total tissue fractions obtained from dorsolateral prefrontal cortex (dlPFC) of 15 MDD and 15 matched non-psychiatric control subjects. A total of 333 miRNAs were reliably detected in the total tissue fraction. Multiple testing following the Benjamini-Hochberg false discovery rate [FDR] showed that 18 miRNAs were significantly altered (1 downregulated 4 up and 13 downregulated; p < 0.05) in MDD subjects. Out of 351 miRNAs reliably expressed in the synaptic fraction, 24 were uniquely expressed at synapse. In addition, 8 miRNAs (miR-215-5p, miR-192-5p, miR-202-5p, miR-19b-3p, miR-423-5p, miR-219a-2-3p; miR-511-5p, miR-483-5p showed significant (FDR corrected; p < 0.05) differential regulation in the synaptic fraction from dlPFC of MDD subjects. In vitro transfection studies and gene ontology revealed involvement of these altered miRNAs in synaptic plasticity, nervous system development, and neurogenesis. A shift in expression ratios (synaptic vs. total fraction) of miR-19b-3p, miR-376c-3p, miR-455-3p, and miR-337-3p were also noted in the MDD group. Moreover, an inverse relationship between the expression of precursor (pre-miR-19b-1, pre-miR-199a-1 and pre-miR-199a-2) and mature (miR-19b-3p, miR-199a-3p) miRNAs was found. Although not significantly, several miRNA processing enzymes (DROSHA [95%], DICER [17%], TARBP2 [38%]) showed increased expression patterns in MDD subjects. Our findings provide new insights into the understanding of the regulation of miRNAs at the synapse and their possible roles in MDD pathogenesis.
Figures



References
-
- Park CS, Gong R, Stuart J, Tang SJ. Molecular network and chromosomal clustering of genes involved in synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:30195–211. - PubMed
-
- Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Disco. 2014;13:622–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

