The role of animals as a source of antimicrobial resistant nontyphoidal Salmonella causing invasive and non-invasive human disease in Vietnam
- PMID: 32920195
- PMCID: PMC7705210
- DOI: 10.1016/j.meegid.2020.104534
The role of animals as a source of antimicrobial resistant nontyphoidal Salmonella causing invasive and non-invasive human disease in Vietnam
Abstract
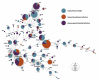
Background: Nontyphoidal Salmonella (NTS) are associated with both diarrhea and bacteremia. Antimicrobial resistance (AMR) is common in NTS in low-middle income countries, but the major source(s) of AMR NTS in humans are not known. Here, we aimed to assess the role of animals as a source of AMR in human NTS infections in Vietnam. We retrospectively combined and analyzed 672 NTS human and animal isolates from four studies in southern Vietnam and compared serovars, sequence types (ST), and AMR profiles. We generated a population structure of circulating organisms and aimed to attribute sources of AMR in NTS causing invasive and noninvasive disease in humans using Bayesian multinomial mixture models.
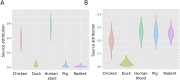
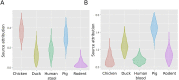
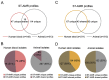
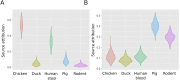
Results: Among 672 NTS isolates, 148 (22%) originated from human blood, 211 (31%) from human stool, and 313 (47%) from animal stool. The distribution of serovars, STs, and AMR profiles differed among sources; serovars Enteritidis, Typhimurium, and Weltevreden were the most common in human blood, human stool, and animals, respectively. We identified an association between the source of NTS and AMR profile; the majority of AMR isolates were isolated from human blood (p < 0.001). Modelling by ST-AMR profile found chickens and pigs were likely the major sources of AMR NTS in human blood and stool, respectively; but unsampled sources were found to be a major contributor.
Conclusions: Antimicrobial use in food animals is hypothesized to play role in the emergence of AMR in human pathogens. Our cross-sectional population-based approach suggests a significant overlap between AMR in NTS in animals and humans, but animal NTS does explain the full extent of AMR in human NTS infections in Vietnam.
Keywords: Antimicrobial resistance; Bacteremia; Diarrhea; Nontyphoidal Salmonella; Zoonosis.
Copyright © 2020 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- An N.Q. Presentation in the 1st GARPS’ Workshop. 2009. Report of antibiotic use in Vietnam.
-
- Angulo F.J., Johnson K.R., Tauxe R.V., Cohen M.L. Origins and consequences of antimicrobial-resistant nontyphoidal Salmonella: implications for the use of fluoroquinolones in food animals. Microb. Drug Resist. 2000;6(1):77–83. - PubMed
-
- Clinical and Laboratory Standards Institute (CLSI) 30th Ed. CLSI; Wayne, PA, USA: 2020. Performance Standards for Antimicrobial Susceptibility Testing: M100.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

