Benzyl and benzoyl benzoic acid inhibitors of bacterial RNA polymerase-sigma factor interaction
- PMID: 32920341
- PMCID: PMC7680358
- DOI: 10.1016/j.ejmech.2020.112671
Benzyl and benzoyl benzoic acid inhibitors of bacterial RNA polymerase-sigma factor interaction
Abstract
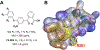
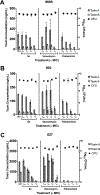
Transcription is an essential biological process in bacteria requiring a core enzyme, RNA polymerase (RNAP). Bacterial RNAP is catalytically active but requires sigma (σ) factors for transcription of natural DNA templates. σ factor binds to RNAP to form a holoenzyme which specifically recognizes a promoter, melts the DNA duplex, and commences RNA synthesis. Inhibiting the binding of σ to RNAP is expected to inhibit bacterial transcription and growth. We previously identified a triaryl hit compound that mimics σ at its major binding site of RNAP, thereby inhibiting the RNAP holoenzyme formation. In this study, we modified this scaffold to provide a series of benzyl and benzoyl benzoic acid derivatives possessing improved antimicrobial activity. A representative compound demonstrated excellent activity against Staphylococcus epidermidis with minimum inhibitory concentrations reduced to 0.5 μg/mL, matching that of vancomycin. The molecular mechanism of inhibition was confirmed using biochemical and cellular assays. Low cytotoxicity and metabolic stability of compounds demonstrated the potential for further studies.
Keywords: Antimicrobial; Bacterial transcription; Inhibitor; RNA polymerase; Sigma factor.
Copyright © 2020 Elsevier Masson SAS. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

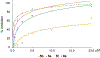


Similar articles
-
Discovery of Antibacterials That Inhibit Bacterial RNA Polymerase Interactions with Sigma Factors.J Med Chem. 2020 Jul 23;63(14):7695-7720. doi: 10.1021/acs.jmedchem.0c00520. Epub 2020 Jul 7. J Med Chem. 2020. PMID: 32633513 Free PMC article.
-
Sulfonamidyl derivatives of sigmacidin: Protein-protein interaction inhibitors targeting bacterial RNA polymerase and sigma factor interaction exhibiting antimicrobial activity against antibiotic-resistant bacteria.Bioorg Chem. 2024 Feb;143:106983. doi: 10.1016/j.bioorg.2023.106983. Epub 2023 Nov 23. Bioorg Chem. 2024. PMID: 38016396
-
The interaction of Bacillus subtilis sigmaA with RNA polymerase.Protein Sci. 2009 Nov;18(11):2287-97. doi: 10.1002/pro.239. Protein Sci. 2009. PMID: 19735077 Free PMC article.
-
Mechanisms for activating bacterial RNA polymerase.FEMS Microbiol Rev. 2010 Sep;34(5):611-27. doi: 10.1111/j.1574-6976.2010.00239.x. Epub 2010 Jun 7. FEMS Microbiol Rev. 2010. PMID: 20629756 Review.
-
Sigma and RNA polymerase: an on-again, off-again relationship?Mol Cell. 2005 Nov 11;20(3):335-45. doi: 10.1016/j.molcel.2005.10.015. Mol Cell. 2005. PMID: 16285916 Review.
Cited by
-
RNA polymerases from low G+C gram-positive bacteria.Transcription. 2021 Aug;12(4):92-102. doi: 10.1080/21541264.2021.1964328. Epub 2021 Aug 17. Transcription. 2021. PMID: 34403307 Free PMC article. Review.
-
Modulators of protein-protein interactions as antimicrobial agents.RSC Chem Biol. 2021 Feb 3;2(2):387-409. doi: 10.1039/d0cb00205d. eCollection 2021 Apr 1. RSC Chem Biol. 2021. PMID: 34458791 Free PMC article. Review.
-
QSAR, Docking, and Molecular Dynamics Simulation Studies of Sigmacidins as Antimicrobials against Streptococci.Int J Mol Sci. 2022 Apr 7;23(8):4085. doi: 10.3390/ijms23084085. Int J Mol Sci. 2022. PMID: 35456906 Free PMC article.
-
Inhibition of bacterial RNA polymerase function and protein-protein interactions: a promising approach for next-generation antibacterial therapeutics.RSC Med Chem. 2024 Mar 26;15(5):1471-1487. doi: 10.1039/d3md00690e. eCollection 2024 May 22. RSC Med Chem. 2024. PMID: 38784472 Free PMC article. Review.
-
Development of a luciferase-based Gram-positive bacterial reporter system for the characterization of antimicrobial agents.Appl Environ Microbiol. 2024 Aug 21;90(8):e0071724. doi: 10.1128/aem.00717-24. Epub 2024 Jul 17. Appl Environ Microbiol. 2024. PMID: 39016615 Free PMC article.
References
-
- O’Neill J, Antimicrobial Resistance: Tackling a Crisis for the Future Health and Wealth of Nations, Review on Antimicrobial Resistance, London, 2014.
-
- Feklistov A, Sharon BD, Darst SA, Gross CA, Bacterial sigma factors: a historical, structural, and genomic perspective, Annu. Rev. Microbiol 68 (2014) 357–376. - PubMed
-
- Borukhov S, Nudler E, RNA polymerase holoenzyme: structure, function and biological implications, Curr. Opin. Microbiol 6 (2003) 93–100. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

