Cell motility and migration as determinants of stem cell efficacy
- PMID: 32920368
- PMCID: PMC7494685
- DOI: 10.1016/j.ebiom.2020.102989
Cell motility and migration as determinants of stem cell efficacy
Abstract
Background: Stem cells` (SC) functional heterogeneity and its poorly understood aetiology impedes clinical development of cell-based therapies in regenerative medicine and oncology. Recent studies suggest a strong correlation between the SC migration potential and their therapeutic efficacy in humans. Designating SC migration as a denominator of functional SC heterogeneity, we sought to identify highly migrating subpopulations within different SC classes and evaluate their therapeutic properties in comparison to the parental non-selected cells.
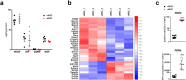
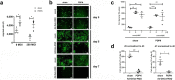
Methods: We selected highly migrating subpopulations from mesenchymal and neural SC (sMSC and sNSC), characterized their features including but not limited to migratory potential, trophic factor release and transcriptomic signature. To assess lesion-targeted migration and therapeutic properties of isolated subpopulations in vivo, surgical transplantation and intranasal administration of MSCs in mouse models of glioblastoma and Alzheimer's disease respectively were performed.
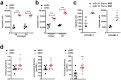
Findings: Comparison of parental non-selected cells with isolated subpopulations revealed superior motility and migratory potential of sMSC and sNSC in vitro. We identified podoplanin as a major regulator of migratory features of sMSC/sNSC. Podoplanin engineering improved oncovirolytic activity of virus-loaded NSC on distantly located glioblastoma cells. Finally, sMSC displayed more targeted migration to the tumour site in a mouse glioblastoma model and remarkably higher potency to reduce pathological hallmarks and memory deficits in transgenic Alzheimer's disease mice.
Interpretation: Functional heterogeneity of SC is associated with their motility and migration potential which can serve as predictors of SC therapeutic efficacy.
Funding: This work was supported in part by the Robert Bosch Stiftung (Stuttgart, Germany) and by the IZEPHA grant.
Keywords: Alzheimer´s disease; Glioblastoma; Intranasal; Mesenchymal stem cells; Neural stem cells; Oncovirolysis.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest Dr. Danielyan reports grants from IZEPHA, during the conduct of the study. Prof.. Schwab, Dr. Schaeffeler and Dr. Winter report grants from Robert Bosch Stiftung (Stuttgart, Germany), during the conduct of the study. Dr. Danielyan, Dr. Schäfer, Prof. Gleiter and Prof. Schwab have a patent US14/634,501, US14/634,484, PCT/EP2016/054055, DE102012107879, EP2888348 pending. All other authors have no competing interests.
Figures





Similar articles
-
Modulating endothelial adhesion and migration impacts stem cell therapies efficacy.EBioMedicine. 2020 Oct;60:102987. doi: 10.1016/j.ebiom.2020.102987. Epub 2020 Sep 14. EBioMedicine. 2020. PMID: 32942121 Free PMC article.
-
A preclinical evaluation of neural stem cell-based cell carrier for targeted antiglioma oncolytic virotherapy.J Natl Cancer Inst. 2013 Jul 3;105(13):968-77. doi: 10.1093/jnci/djt141. J Natl Cancer Inst. 2013. PMID: 23821758 Free PMC article.
-
HuCNS-SC Human NSCs Fail to Differentiate, Form Ectopic Clusters, and Provide No Cognitive Benefits in a Transgenic Model of Alzheimer's Disease.Stem Cell Reports. 2017 Feb 14;8(2):235-248. doi: 10.1016/j.stemcr.2016.12.019. Stem Cell Reports. 2017. PMID: 28199828 Free PMC article.
-
Migration and fate of therapeutic stem cells in different brain disease models.Neuroscience. 2011 Dec 1;197:37-47. doi: 10.1016/j.neuroscience.2011.08.063. Epub 2011 Sep 14. Neuroscience. 2011. PMID: 21946010 Free PMC article. Review.
-
Therapeutic potential of neurogenesis for prevention and recovery from Alzheimer's disease: allopregnanolone as a proof of concept neurogenic agent.Curr Alzheimer Res. 2006 Jul;3(3):185-90. doi: 10.2174/156720506777632817. Curr Alzheimer Res. 2006. PMID: 16842093 Review.
Cited by
-
More Human BM-MSC With Similar Subpopulation Composition and Functional Characteristics Can Be Produced With a GMP-Compatible Fabric Filter System Compared to Density Gradient Technique.Front Cell Dev Biol. 2021 Mar 29;9:638798. doi: 10.3389/fcell.2021.638798. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33869188 Free PMC article.
-
Small volume bone marrow aspirates with high progenitor cell concentrations maximize cell therapy dose manufacture and substantially reduce donor hemoglobin loss.BMC Med. 2023 Sep 19;21(1):360. doi: 10.1186/s12916-023-03059-3. BMC Med. 2023. PMID: 37726769 Free PMC article.
-
Emerging delivery strategy for oncolytic virotherapy.Mol Ther Oncol. 2024 Apr 29;32(2):200809. doi: 10.1016/j.omton.2024.200809. eCollection 2024 Jun 20. Mol Ther Oncol. 2024. PMID: 38845744 Free PMC article. Review.
-
Sensory nerves drive migration of dental pulp stem cells via the CGRP-Ramp1 axis in pulp repair.Cell Mol Life Sci. 2024 Aug 28;81(1):373. doi: 10.1007/s00018-024-05400-2. Cell Mol Life Sci. 2024. PMID: 39196292 Free PMC article.
-
Advances in stem cell therapy in Alzheimer's disease: a comprehensive clinical trial review.Stem Cell Investig. 2022 Feb 21;9:2. doi: 10.21037/sci-2021-063. eCollection 2022. Stem Cell Investig. 2022. PMID: 35280344 Free PMC article. Review.
References
-
- Mendonça LS, Onofre I, Miranda CO, Perfeito R, Nóbrega C, de Almeida LP. Stem cell-based therapies for polyglutamine diseases. Adv. Exp. Med. Biol. 2018;1049:439–466. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

